








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 121-130.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1516
Previous Articles Next Articles
FAN Ya-peng( ), RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei(
), RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei( )
)
Received:2020-12-15
Online:2021-08-26
Published:2021-09-10
Contact:
YE Wu-wei
E-mail:fanyp0605@126.com;yew158@163.com
FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum[J]. Biotechnology Bulletin, 2021, 37(8): 121-130.
| 引物名称Primer name | 引物序列Primers sequence(5'-3') | 描述Description |
|---|---|---|
| GhZAT12 | F:ATGAAGAGAGGCAGAGATATTGATG | GhZAT12基因引物 Primer pairs for GhZAT12 gene |
| R:CTATATAAAACAATGAACAACCGGA | ||
| GFP-GhZAT12 | F:AGAACACGGGGGACTCTAGAATGAAGAGAGGCAGAGATATTG | GhZAT12亚细胞定位载体引物 Primer pairs for subcellular localization vector |
| R:TAGTCAGGCGCGCCGGTACCTATAAAACAATGAACAACCGG | ||
| Actin(AY305733) | F:ATCCTCCGTCTTGACCTTG | 内参基因引物 Primer pairs for internal reference gene |
| R:TGTCCGTCAGGCAACTCAT | ||
| QGhZAT12 | F:CTGCTCTGAATGACGGGTTG | GhZAT12荧光定量引物 Primer pairs for qRT-PCR of GhZAT12 |
| R:CCGGAGTTGGTGTCACTTTC |
Table 1 Primer pairs used in the experiments
| 引物名称Primer name | 引物序列Primers sequence(5'-3') | 描述Description |
|---|---|---|
| GhZAT12 | F:ATGAAGAGAGGCAGAGATATTGATG | GhZAT12基因引物 Primer pairs for GhZAT12 gene |
| R:CTATATAAAACAATGAACAACCGGA | ||
| GFP-GhZAT12 | F:AGAACACGGGGGACTCTAGAATGAAGAGAGGCAGAGATATTG | GhZAT12亚细胞定位载体引物 Primer pairs for subcellular localization vector |
| R:TAGTCAGGCGCGCCGGTACCTATAAAACAATGAACAACCGG | ||
| Actin(AY305733) | F:ATCCTCCGTCTTGACCTTG | 内参基因引物 Primer pairs for internal reference gene |
| R:TGTCCGTCAGGCAACTCAT | ||
| QGhZAT12 | F:CTGCTCTGAATGACGGGTTG | GhZAT12荧光定量引物 Primer pairs for qRT-PCR of GhZAT12 |
| R:CCGGAGTTGGTGTCACTTTC |
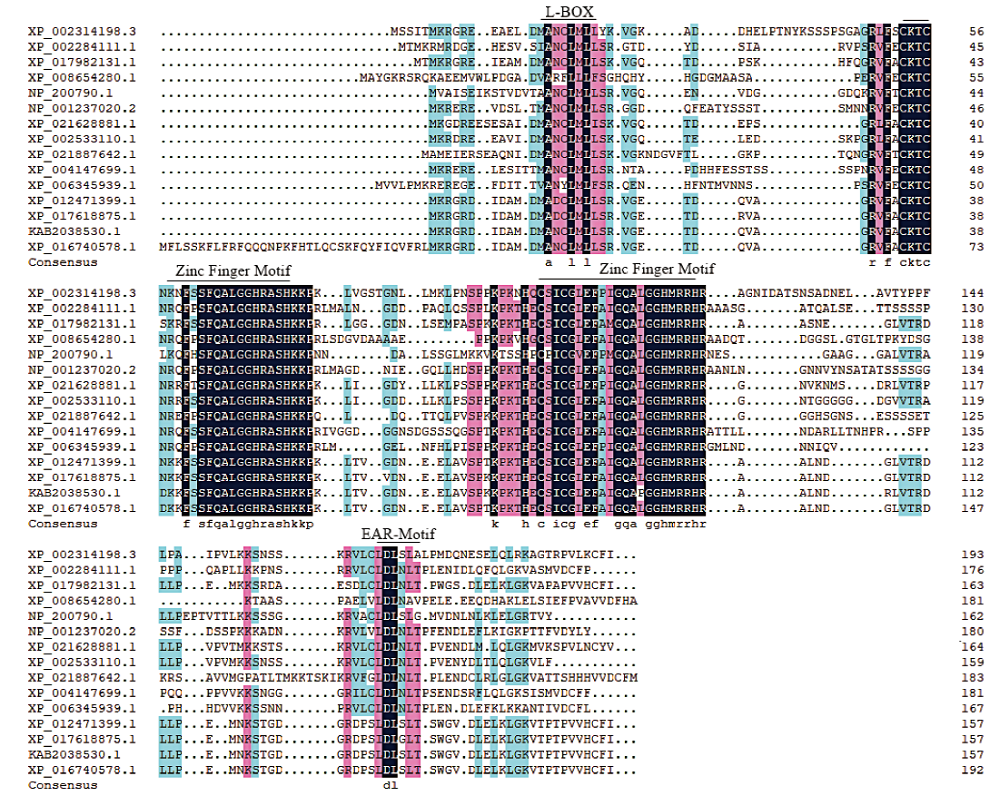
Fig.2 Multiple sequence alignment of homologous amino acids in GhZAT12 protein XP_002314198.3 Populus trichocarpa, XP_002284111.1 Vitis vinifera, XP_017982131.1 Theobroma cacao, XP_008654280.1 Zea mays, NP_200790.1 Arabidopsis thaliana, NP_001237020.2 Glycine max, XP_021628881.1 Manihot esculenta, XP_002533110.1 Ricinus communis, XP_021887642.1 Carica papaya, XP_004147699.1 Cucumis sativus, XP_006345939.1 Solanum tuberosum, XP_012471399.1 Gossypium raimondii, XP_017618875.1 Gossypium arboretum, KAB2038530.1 Gossypium barbadense, XP_016740578.1 Gossypium hirsutum
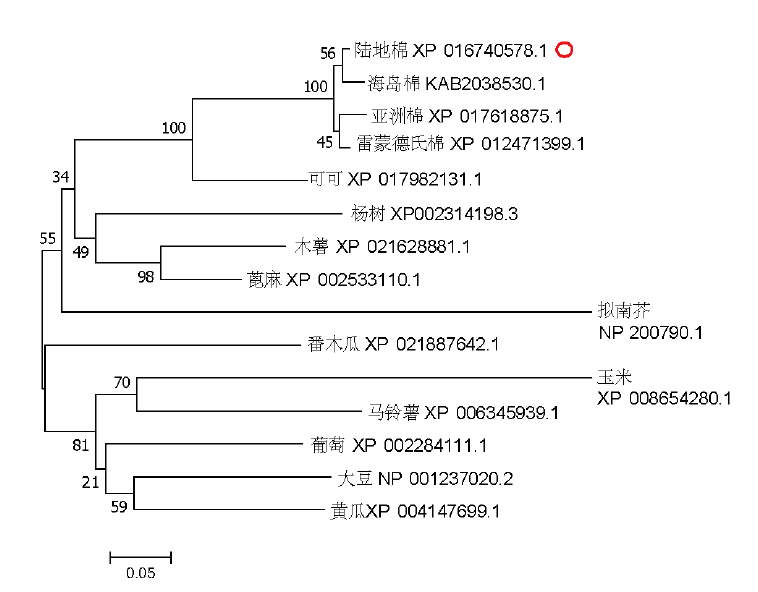
Fig.3 Phylogenetic tree of ZAT12 homologous proteins in different species Parameters are set to be neighbor-joining,boostrap value=1 000. The numbers on the tree branches represent bootstrap confidence values as “Bootstrap” is 1000

Fig.10 Subcellular localization of GhZAT12 in tobacco leaves a,d:Confocal section of tobacco leave expressing GFP and GhZAT12-GFP,respectively. b,e:Bright field. c,f:Fluorescence and bright field merged image. Bar=50 μm
| [1] |
Rocha-Munive MG, Soberón M, Castañeda S, et al. Evaluation of the impact of genetically modified cotton after 20 years of cultivation in Mexico[J]. Frontiers in Bioengineering and Biotechnology, 2018, 6:82.
doi: 10.3389/fbioe.2018.00082 pmid: 29988354 |
| [2] |
Jin S, Xu C, Li G, et al. Functional characterization of a type 2 metallothionein gene, SsMT2, from alkaline-tolerant Suaeda salsa[J]. Scientific Reports, 2017, 7(1)17914.
doi: 10.1038/s41598-017-18263-4 URL |
| [3] |
Wang Y, Ma H, Liu G, et al. Analysis of gene expression profile of limonium bicolor under NaHCO3 stress using cDNA microarray[J]. Plant Molecular Biology Reporter, 2008, 26(3):241-254.
doi: 10.1007/s11105-008-0037-4 URL |
| [4] |
Sun Z, Liu R, Guo B, et al. Ectopic expression of GmZAT4, a puta-tive C2H2-type zinc finger protein, enhances PEG and NaCl stress tolerances in Arabidopsis thaliana[J]. 3 Biotech, 2019, 9(5):166.
doi: 10.1007/s13205-019-1673-0 URL |
| [5] |
Meissner R, Michael AJ. Isolation and characterisation of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs[J]. Plant Molecular Biology, 1997, 33(4):615-624.
pmid: 9132053 |
| [6] |
Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants[J]. Cell Mol Life Sci, 2008, 65(7-8):1150-1160.
doi: 10.1007/s00018-007-7473-4 pmid: 18193167 |
| [7] |
Takatsuji H. Zinc-finger proteins:the classical zinc finger emerges in contemporary plant science[J]. Plant Molecular Biology, 1999, 39(6):1073-1078.
pmid: 10380795 |
| [8] |
Zhang H, Liu Y, Wen F, et al. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice[J]. J Exp Bot, 2014, 65(20):5795-5809.
doi: 10.1093/jxb/eru313 pmid: 25071223 |
| [9] |
Zhang H, Ni L, Liu Y, et al. The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice[J]. J Integr Plant Biol, 2012, 54(7):500-510.
doi: 10.1111/j.1744-7909.2012.01135.x |
| [10] |
Huang F, Chi Y, Meng Q, et al. GmZFP1 encoding a single zinc finger protein is expressed with enhancement in reproductive organs and late seed development in soybean(Glycine max)[J]. Mol Biol Rep, 2006, 33(4):279-285.
pmid: 17077988 |
| [11] |
Yu GH, Jiang LL, Ma XF, et al. A soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic Arabidopsis[J]. PLoS One, 2014, 9(10):e109399.
doi: 10.1371/journal.pone.0109399 URL |
| [12] | Zhang D, Tong J, Xu Z, et al. Soybean C2H2-type zinc finger protein GmZFP3 with conserved QALGGH motif negatively regulates drought responses in transgenic Arabidopsis[J]. Frontiers in Plant Science, 2016, 7:325. |
| [13] |
Sakamoto H, Maruyama K, Sakuma Y, et al. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions[J]. Plant Physiology, 2004, 136(1):2734-2746.
pmid: 15333755 |
| [14] |
Li G, Tai FJ, Zheng Y, et al. Two cotton Cys2/His2-type zinc-finger proteins, GhDi19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling[J]. Plant Molecular Biology, 2010, 74(4-5):437-452.
doi: 10.1007/s11103-010-9684-6 URL |
| [15] | An YM, Song LL, Liu YR, et al. De novo transcriptional analysis of alfalfa in response to saline-alkaline stress[J]. Frontiers in Plant science, 2016, 7:931. |
| [16] |
Le CTT, Brumbarova T, Ivanov R, et al. Zinc finger of Arabidopsis thaliana12(ZAT12)interacts with FER-like iron deficiency-induced transcription factor(FIT)linking iron deficiency and oxidative stress responses[J]. Plant Physiology, 2016, 170(1):540-557.
doi: 10.1104/pp.15.01589 URL |
| [17] |
Rizhsky L, Davletova S, Liang H, et al. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis[J]. Journal of Biological Chemistry, 2004, 279(12):11736-11743.
doi: 10.1074/jbc.M313350200 URL |
| [18] |
Davletova S, Schlauch K, Coutu J, et al. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis[J]. Plant Physiology, 2005, 139(2):847-856.
pmid: 16183833 |
| [19] |
Rai AC, Singh M, Shah K. Engineering drought tolerant tomato plants over-expressing BcZAT12 gene encoding a C2H2 zinc finger transcription factor[J]. Phytochemistry, 2013, 85:44-50.
doi: 10.1016/j.phytochem.2012.09.007 URL |
| [20] |
Wang M, Yuan J, Qin L, et al. TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging[J]. Plant Biotechnology Journal, 2020, 18(3):791-804.
doi: 10.1111/pbi.v18.3 URL |
| [21] |
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C T method[J]. Nature Protocols, 2008, 3(6):1101.
pmid: 18546601 |
| [22] | 张新宇, 林书岱, 张涛, 等. 棉花C2H2类型锌指蛋白基因GhSIZ1的克隆及表达分析[J]. 棉花学报, 2015, 27(3):189-197. |
| Zhang XY, lin SD, Zhang T, et al. Cloning and expression analysis of GhSIZ1, encoding a C2H2 zinc finger protein in cotton(Gossypium hirsutum)[J]. Cotton Science, 2015, 27(3):189-197. | |
| [23] |
Li F, Fan G, Lu C, et al. Genome sequence of cultivated Upland cotton(Gossypium hirsutum TM-1)provides insights into genome evolution[J]. Nature Biotechnology, 2015, 33(5):524-530.
doi: 10.1038/nbt.3208 URL |
| [24] |
Ullah A, Qamar MTU, Nisar M, et al. Characterization of a novel cotton MYB gene, GhMYB108-like responsive to abiotic stresses[J]. Molecular Biology Reports, 2020, 47(3):1573-1581.
doi: 10.1007/s11033-020-05244-6 pmid: 31933260 |
| [25] |
Wang X, Lu X, Malik WA, et al. Differentially expressed bZIP transcription factors confer multi-tolerances in Gossypium hirsutum L.[J]. International Journal of Biological Macromolecules, 2020, 146:569-578.
doi: 10.1016/j.ijbiomac.2020.01.013 URL |
| [26] |
Gu L, Wang H, Wei H, et al. Identification, expression, and functional analysis of the group IId WRKY subfamily in upland cotton(Gossypium hirsutum L. )[J]. Frontiers in Plant Science, 2018, 9:1684.
doi: 10.3389/fpls.2018.01684 URL |
| [27] |
Gu L, Wei H, Wang H, et al. Characterization and functional analy-sis of GhWRKY42, a group IId WRKY gene, in upland cotton(Gos-sypium hirsutum L. )[J]. BMC Genetics, 2018, 19(1):48.
doi: 10.1186/s12863-018-0653-4 URL |
| [28] |
Xiong XP, Sun SC, Zhang XY, et al. GhWRKY70D13 regulates resistance to verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways[J]. Frontiers in Plant Science, 2020, 11:69.
doi: 10.3389/fpls.2020.00069 URL |
| [29] |
Kundu A, Das S, Basu S, et al. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for Aluminum and proton stress tolerance and lateral root initiation in cotton[J]. Plant Biology, 2019, 21(1):35-44.
doi: 10.1111/plb.12895 pmid: 30098101 |
| [30] |
周垚均, 任艳萍, 代培红, 等. 棉花四个C2H2锌指蛋白基因的克隆与表达分析[J]. 分子植物育种, 2020. DOI: http://kns.chki.net/kcms/detail/46.1068.S.20200522.1131.002.html.
doi: http://kns.chki.net/kcms/detail/46.1068.S.20200522.1131.002.html |
|
Zhou JY, Ren YP, Dai PH, et al. Cloning and Expression Analysis of Four C2H2 Zinc Protein Gene in Cotton(Gossypium hirsutum L.)[J]. Molecular Plant Breeding, 2020. DOI: http://kns.chki.net/kcms/detail/46.1068.S.20200522.1131.002.html.
doi: http://kns.chki.net/kcms/detail/46.1068.S.20200522.1131.002.html |
|
| [31] |
Kubo K, Sakamoto A, Kobayashi A, et al. Cys2/His2 zinc-finger protein family of petunia:evolution and general mechanism of target-sequence recognition[J]. Nucleic Acids Research, 1998, 26(2):608-615.
pmid: 9421523 |
| [32] |
Ohta M, Matsui K, Hiratsu K, et al. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression[J]. The Plant Cell, 2001, 13(8):1959-1968.
doi: 10.1105/TPC.010127 URL |
| [33] |
Yamaji N, Huang CF, Nagao S, et al. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice[J]. The Plant Cell, 2009, 21(10):3339-3349.
doi: 10.1105/tpc.109.070771 pmid: 19880795 |
| [34] |
Xu SM, Wang XC, Chen J. Zinc finger protein 1(ThZF1)from salt cress(Thellungiella halophila)is a Cys-2/His-2-type transcription factor involved in drought and salt stress[J]. Plant Cell Reports, 2007, 26(4):497-506.
doi: 10.1007/s00299-006-0248-9 URL |
| [35] |
Zhang H, Gao X, Zhi Y, et al. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato[J]. New Phytologist, 2019, 223(4):1918-1936.
doi: 10.1111/nph.15925 pmid: 31091337 |
| [36] | An Y, Yang XX, Zhang L, et al. Alfalfa MsCBL4 enhances calcium metabolism but not sodium transport in transgenic tobacco under salt and saline-alkali stress[J]. Plant Cell Reports, 2020:1-15. |
| [37] |
Yang Y, Wu Y, Ma L, et al. The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane H+-ATPase activity and promotes alkali tolerance in Arabidopsis[J]. The Plant Cell, 2019, 31(6):1367-1384.
doi: 10.1105/tpc.18.00568 URL |
| [38] | Zhang B, Chen X, Lu X, et al. Transcriptome analysis of Gossypium hirsutum L. reveals different mechanisms among NaCl, NaOH and Na2CO3 stress tolerance[J]. Scientific Reports, 2018, 8(1):1-14. |
| [39] | 陈宗新, 陈俊飞, 王亚琴. 水稻C2H2型锌指蛋白OsZAT12转化拟南芥功能的初步研究[J]. 华南师范大学学报:自然科学版, 2019, 51(1):63-69. |
| Chen ZX, Chen JF, Wang YQ. A Preliminary study of OsZAT12 with C2H2-type zinc finger transforming Arabidopsis[J]. Journal of South China Normal University:Natural Science Edition, 2019, 51(1):63-69. |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [3] | WANG Yi-fan, HOU Lin-hui, CHANG Yong-chun, YANG Ya-jie, CHEN Tian, ZHAO Zhu-yue, RONG Er-hua, WU Yu-xiang. Synthesis and Character Identification of Allohexaploid Between Gossypium hirsutum and G. gossypioides [J]. Biotechnology Bulletin, 2023, 39(5): 168-176. |
| [4] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [5] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [6] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [7] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [8] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| [9] | CHEN Jia-min, LIU Yong-jie, MA Jin-xiu, LI Dan, GONG Jie, ZHAO Chang-ping, GENG Hong-wei, GAO Shi-qing. Expression Pattern Analysis of Histone Methyltransferase Under Drought Stress in Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(7): 51-61. |
| [10] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| [11] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [12] | WANG Nan, ZHANG Rui, PAN Yang-yang, HE Hong-hong, WANG Jing-lei, CUI Yan, YU Si-jiu. Cloning of Bos grunniens TGF-β1 Gene and Its Expression in Major Organs of Female Reproductive System [J]. Biotechnology Bulletin, 2022, 38(6): 279-290. |
| [13] | LI Yu-hang, WANG Xing-ping, YANG Jian, LUORENG Zhuo-ma, REN Qian-qian, WEI Da-wei, MA Yun. Expression and Functional Analysis of miR-665 in Bovine Mammary Epithelial Cell Inflammation [J]. Biotechnology Bulletin, 2022, 38(5): 159-168. |
| [14] | SUN Bao-zhen, QUAN Long-ping, KANG Hui, YAO Yu-xin, SHEN Tian, CHEN Wei-ping, DU Yuan-peng, GAO Zhen. Back-splicing Primers-based PCR Method for Specific Detection of circRNA [J]. Biotechnology Bulletin, 2022, 38(5): 279-285. |
| [15] | LI Yang, ZHANG Xiao-tian, PIAO Jing-zi, ZHOU Ru-jun, LI Zi-bo, GUAN Hai-wen. Cloning and Bioinformatics Analysis of Blue-light Receptor EaWC 1 Gene in Elsinoë arachidis [J]. Biotechnology Bulletin, 2022, 38(5): 93-99. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||