








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 142-151.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0975
Previous Articles Next Articles
LI Zhi-qi( ), YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin(
), YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin( )
)
Received:2022-08-10
Online:2023-05-26
Published:2023-06-08
Contact:
ZHANG Ai-qin
E-mail:lzq1321059957@163.com;aiqinaegean@nefu.edu.cn
LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes[J]. Biotechnology Bulletin, 2023, 39(5): 142-151.
| 基因名称 Gene name | 引物方向 Primer direction | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| AtACTIN | Forward | CATCAGGAAGGACTTGTACGG |
| Reverse | GATGGACCTGACTCGTCATAC | |
| AtSNAT1 | Forward | CGTCTATTAGCCCTCCTCC |
| Reverse | GCCATCCCACCTTATCACA | |
| AtSNAT2 | Forward | GGCACAATCTCCACTCCTC |
| Reverse | TCAACCACAACATCCCATA | |
| AtASMT | Forward | GGTCAACGGTTCAACTCCA |
| Reverse | ATCGCCCTCAACATTCTCA | |
| AtCOMT | Forward | ATGCTCCTTCTCATCCTGG |
| Reverse | CACGCAATGTTCGTCACTC | |
| EsACTIN | Forward | GCACAATCCAAAAGAGGTATTCTCACCT |
| Reverse | GGAGCCTCGGTAAGAAGAACAGGG | |
| EsSNAT1 | Forward | AAGAATAACAATACGACCCA |
| Reverse | ACATCAATCTCACCACCAG | |
| EsASMT | Forward | ATGTTACCGAAGTTGCG |
| Reverse | CGTTCTTTGCCTGTGCT | |
| EsCOMT | Forward | CGGCTTACTCCGTCCTCAC |
| Reverse | TAGCACCAATGCCACCACC |
Table 1 Primer sequences used for RT-qPCR analysis
| 基因名称 Gene name | 引物方向 Primer direction | 引物序列 Primer sequence(5'-3') |
|---|---|---|
| AtACTIN | Forward | CATCAGGAAGGACTTGTACGG |
| Reverse | GATGGACCTGACTCGTCATAC | |
| AtSNAT1 | Forward | CGTCTATTAGCCCTCCTCC |
| Reverse | GCCATCCCACCTTATCACA | |
| AtSNAT2 | Forward | GGCACAATCTCCACTCCTC |
| Reverse | TCAACCACAACATCCCATA | |
| AtASMT | Forward | GGTCAACGGTTCAACTCCA |
| Reverse | ATCGCCCTCAACATTCTCA | |
| AtCOMT | Forward | ATGCTCCTTCTCATCCTGG |
| Reverse | CACGCAATGTTCGTCACTC | |
| EsACTIN | Forward | GCACAATCCAAAAGAGGTATTCTCACCT |
| Reverse | GGAGCCTCGGTAAGAAGAACAGGG | |
| EsSNAT1 | Forward | AAGAATAACAATACGACCCA |
| Reverse | ACATCAATCTCACCACCAG | |
| EsASMT | Forward | ATGTTACCGAAGTTGCG |
| Reverse | CGTTCTTTGCCTGTGCT | |
| EsCOMT | Forward | CGGCTTACTCCGTCCTCAC |
| Reverse | TAGCACCAATGCCACCACC |
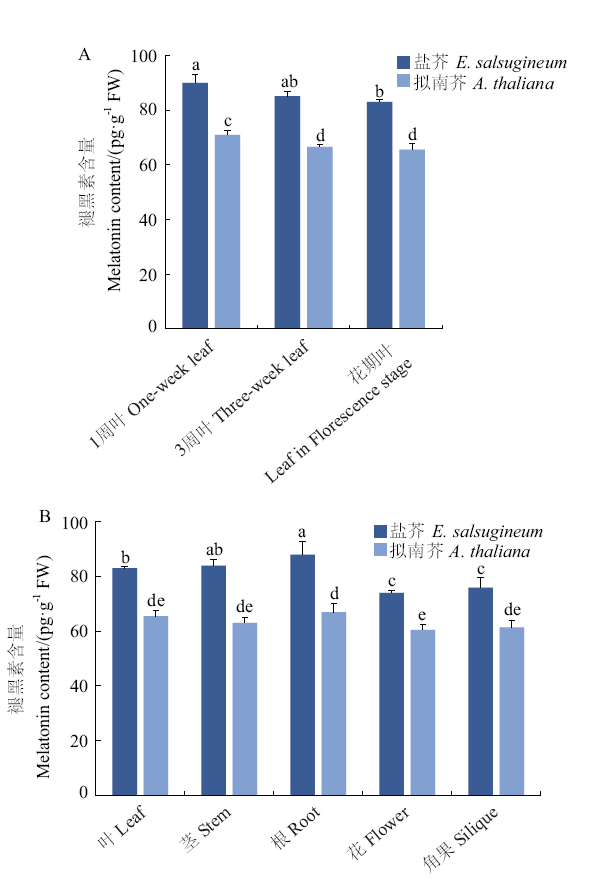
Fig. 1 Melatonin contents in different tissues of E. salsug-ineum and A. thaliana A: Vegetative growth stage. B: Reproductive growth stage. Different letters indicate significant difference(P < 0.05). The same below

Fig. 3 Multiple alignment of amino acid sequences of melatonin synthases in different plants Black boxes indicate conserved functional sites or domains in proteases, and * indicates the binding sites of phenolic substrate. At: Arabidopsis thaliana; Es: Eutrema salsugineum; Vv: Vitis vinifera L.; Sl: Solanum lycopersicum L.; Os: Oryza sativa L.; Zm: Zea mays L. The same below
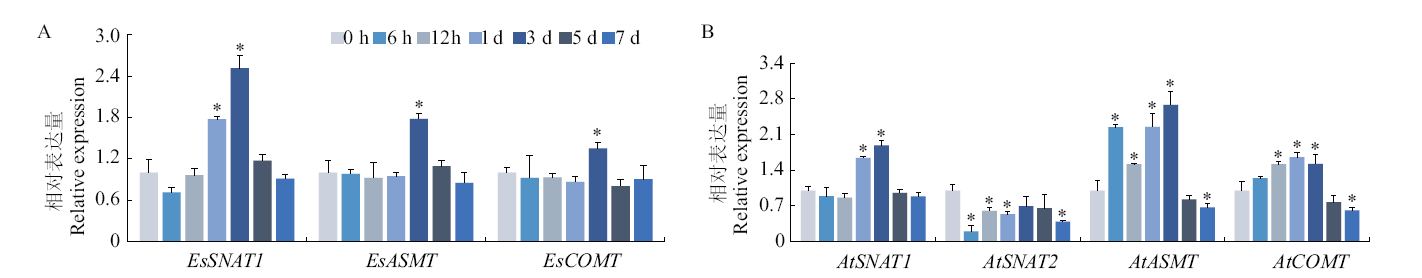
Fig. 5 Expression pattern of melatonin synthesis genes in response to salt stress A: E. salsugineum. B: A. thaliana. * indicates significant difference between untreated and salt treated plants at different time point(P < 0.05)

Fig. 6 Salt-responsive phenotypes of E. salsugineum and A. thaliana treated with exogenous melatonin A: E. salsugineum. B: A. thaliana. C: Control. MT: Melatonin treatment. S: Salt treatment. S+MT: Salt and melatonin treatment
| [1] |
Morton MJL, Awlia M, Al-Tamimi N, et al. Salt stress under the scalpel - dissecting the genetics of salt tolerance[J]. Plant J, 2019, 97(1): 148-163.
doi: 10.1111/tpj.2019.97.issue-1 URL |
| [2] |
Gong ZZ, Xiong LM, Shi HZ, et al. Plant abiotic stress response and nutrient use efficiency[J]. Sci China Life Sci, 2020, 63(5): 635-674.
doi: 10.1007/s11427-020-1683-x pmid: 32246404 |
| [3] |
Parihar P, Singh S, Singh R, et al. Effect of salinity stress on plants and its tolerance strategies: a review[J]. Environ Sci Pollut Res Int, 2015, 22(6): 4056-4075.
doi: 10.1007/s11356-014-3739-1 URL |
| [4] |
Apse MP, Blumwald E. Engineering salt tolerance in plants[J]. Curr Opin Biotechnol, 2002, 13(2): 146-150.
doi: 10.1016/S0958-1669(02)00298-7 URL |
| [5] |
Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324.
doi: 10.1016/j.cell.2016.08.029 URL |
| [6] |
Park HJ, Kim WY, Yun DJ. A new insight of salt stress signaling in plant[J]. Mol Cells, 2016, 39(6): 447-459.
doi: 10.14348/molcells.2016.0083 pmid: 27239814 |
| [7] |
Kanwar MK, Yu JQ, Zhou J. Phytomelatonin: Recent advances and future prospects[J]. J Pineal Res, 2018, 65(4): e12526.
doi: 10.1111/jpi.2018.65.issue-4 URL |
| [8] | Lerner AB, Case JD, Takahashi Y, et al. Isolation of melatonin, the pineal gland factor that lightens melanocytes[J]. J Am Chem Soc, 1958, 80(10): 2587. |
| [9] |
Dubbels R, Reiter RJ, Klenke E, et al. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry[J]. J Pineal Res, 1995, 18(1): 28-31.
doi: 10.1111/j.1600-079x.1995.tb00136.x pmid: 7776176 |
| [10] |
Hattori A, Migitaka H, Iigo M, et al. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates[J]. Biochem Mol Biol Int, 1995, 35(3): 627-634.
pmid: 7773197 |
| [11] |
Lee HY, Byeon Y, Lee K, et al. Cloning of Arabidopsis serotonin N-acetyltransferase and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations[J]. J Pineal Res, 2014, 57(4): 418-426.
doi: 10.1111/jpi.12181 URL |
| [12] | 宋雪飞. 褪黑素调控番茄幼苗耐盐性的浓度效应及其生理机制的研究[D]. 扬州: 扬州大学, 2018. |
| Song XF. Study on the concentration effect of melatonin on salt tolerance of tomato seedlings and its physiological mechanism[D]. Yangzhou: Yangzhou University, 2018. | |
| [13] | 乔沛, 殷菲胧, 王雨萱, 等. 外源褪黑素处理对采后荔枝褐变及活性氧代谢的影响[J]. 食品工业科技, 2021, 42(6): 282-287. |
| Qiao P, Yin FL, Wang YX, et al. Effects of exogenous melatonin on browning and active oxygen metabolism of postharvest Litchi[J]. Sci Technol Food Ind, 2021, 42(6): 282-287. | |
| [14] |
Sun CL, Liu LJ, Wang LX, et al. Melatonin: a master regulator of plant development and stress responses[J]. J Integr Plant Biol, 2021, 63(1): 126-145.
doi: 10.1111/jipb.12993 |
| [15] |
Zhan HS, Nie XJ, Zhang T, et al. Melatonin: a small molecule but important for salt stress tolerance in plants[J]. Int J Mol Sci, 2019, 20(3): 709.
doi: 10.3390/ijms20030709 URL |
| [16] |
Li JP, Liu J, Zhu TT, et al. The role of melatonin in salt stress responses[J]. Int J Mol Sci, 2019, 20(7): 1735.
doi: 10.3390/ijms20071735 URL |
| [17] |
Gong QQ, Li PH, Ma SS, et al. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana[J]. Plant J, 2005, 44(5): 826-839.
doi: 10.1111/tpj.2005.44.issue-5 URL |
| [18] |
Amtmann A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants[J]. Mol Plant, 2009, 2(1): 3-12.
doi: 10.1093/mp/ssn094 pmid: 19529830 |
| [19] |
Inan G, Zhang Q, Li PH, et al. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles[J]. Plant Physiol, 2004, 135(3): 1718-1737.
doi: 10.1104/pp.104.041723 URL |
| [20] |
Pelagio-Flores R, Muñoz-Parra E, Ortiz-Castro R, et al. Melatonin regulates Arabidopsis root system architecture likely acting independently of auxin signaling[J]. J Pineal Res, 2012, 53(3): 279-288.
doi: 10.1111/j.1600-079X.2012.00996.x pmid: 22507071 |
| [21] | 张悦新. 褪黑素对棉花耐盐性的影响及相关基因GhSNAT34的功能研究[D]. 郑州: 郑州大学, 2021. |
| Zhang YX. Study on effect of melatonin on salt tolerance of cotton and function of GhSNAT34 gene[D]. Zhengzhou: Zhengzhou University, 2021. | |
| [22] |
Li JH, Yang YQ, Sun K, et al. Exogenous melatonin enhances cold, salt and drought stress tolerance by improving antioxidant defense in tea plant(Camellia sinensis(L.) O. Kuntze)[J]. Molecules, 2019, 24(9): 1826.
doi: 10.3390/molecules24091826 URL |
| [23] | 吴艳迪. 葡萄褪黑素含量变化及其合成基因SNAT原核表达分析[D]. 北京: 中国农业科学院, 2018. |
| Wu YD. Changes of melatonin content and prokaryotic expression analysis of SNAT gene in grapevine[D]. Beijing: Chinese Academy of Agricultural Sciences, 2018. | |
| [24] |
Arnao MB, Hernández-Ruiz J. Chemical stress by different agents affects the melatonin content of barley roots[J]. J Pineal Res, 2009, 46(3): 295-299.
doi: 10.1111/j.1600-079X.2008.00660.x pmid: 19196434 |
| [25] |
Byeon Y, Lee HY, Lee K, et al. Cellular localization and kinetics of the rice melatonin biosynthetic enzymes SNAT and ASMT[J]. J Pineal Res, 2014, 56(1): 107-114.
doi: 10.1111/jpi.12103 pmid: 24134674 |
| [26] |
Liu DD, Sun XS, Liu L, et al. Overexpression of the melatonin synthesis-related gene SlCOMT1 improves the resistance of tomato to salt stress[J]. Molecules, 2019, 24(8): 1514.
doi: 10.3390/molecules24081514 URL |
| [27] |
Chen YE, Mao JJ, Sun LQ, et al. Exogenous melatonin enhances salt stress tolerance in maize seedlings by improving antioxidant and photosynthetic capacity[J]. Physiol Plant, 2018, 164(3): 349-363.
doi: 10.1111/ppl.2018.164.issue-3 URL |
| [28] |
Yan FY, Zhang JY, Li WW, et al. Exogenous melatonin alleviates salt stress by improving leaf photosynthesis in rice seedlings[J]. Plant Physiol Biochem, 2021, 163: 367-375.
doi: 10.1016/j.plaphy.2021.03.058 URL |
| [29] |
Zhang ZH, Liu LT, Li HY, et al. Exogenous melatonin promotes the salt tolerance by removing active oxygen and maintaining ion balance in wheat(Triticum aestivum L.)[J]. Front Plant Sci, 2022, 12: 787062.
doi: 10.3389/fpls.2021.787062 URL |
| [30] |
Xu LL, Xiang GQ, Sun QH, et al. Melatonin enhances salt tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines[J]. Hortic Res, 2019, 6: 114.
doi: 10.1038/s41438-019-0197-4 |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | KANG Ling-yun, HAN Lu-lu, HAN De-ping, CHEN Jian-sheng, GAN Han-ling, XING Kai, MA You-ji, CUI Kai. Effect of Melatonin on Protecting the Jejunum Mucosal Epithelial Cells from Oxidative Stress Damage [J]. Biotechnology Bulletin, 2023, 39(9): 291-299. |
| [3] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [4] | HAN Zhi-yang, JIA Zi-miao, LIANG Qiu-ju, WANG Ke, TANG Hua-li, YE Xing-guo, ZHANG Shuang-xi. Salt Tolerance at Seedling Stage and Analysis of Selenium and Folic Acid Content in Seeds in Two Sets of Wheat-Dasypyrum villosum Chromosom Additional Lines [J]. Biotechnology Bulletin, 2023, 39(8): 185-193. |
| [5] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [6] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [7] | LIU Kui, LI Xing-fen, YANG Pei-xin, ZHONG Zhao-chen, CAO Yi-bo, ZHANG Ling-yun. Functional Study and Validation of Transcriptional Coactivator PwMBF1c in Picea wilsonii [J]. Biotechnology Bulletin, 2023, 39(5): 205-216. |
| [8] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [9] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [10] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [11] | YAO Xiao-wen, LIANG Xiao, CHEN Qing, WU Chun-ling, LIU Ying, LIU Xiao-qiang, SHUI Jun, QIAO Yang, MAO Yi-ming, CHEN Yin-hua, ZHANG Yin-dong. Study on the Expression Pattern of Genes in Lignin Biosynthesis Pathway of Cassava Resisting to Tetranychus urticae [J]. Biotechnology Bulletin, 2023, 39(2): 161-171. |
| [12] | LI Yan-xia, WANG Jin-peng, FENG Fen, BAO Bin-wu, DONG Yi-wen, WANG Xing-ping, LUORENG Zhuo-ma. Effects of Escherichia coli Dairy Cow Mastitis on the Expressions of Milk-producing Trait Related Genes [J]. Biotechnology Bulletin, 2023, 39(2): 274-282. |
| [13] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| [14] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [15] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||