








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 174-185.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0149
Previous Articles Next Articles
LI Yu-qing( ), WU Nan, LUO Jian-rang(
), WU Nan, LUO Jian-rang( )
)
Received:2024-02-07
Online:2024-08-26
Published:2024-06-26
Contact:
LUO Jian-rang
E-mail:liyuqing@nwafu.edu.cn;luojianrang@nwafu.edu.cn
LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui[J]. Biotechnology Bulletin, 2024, 40(8): 174-185.
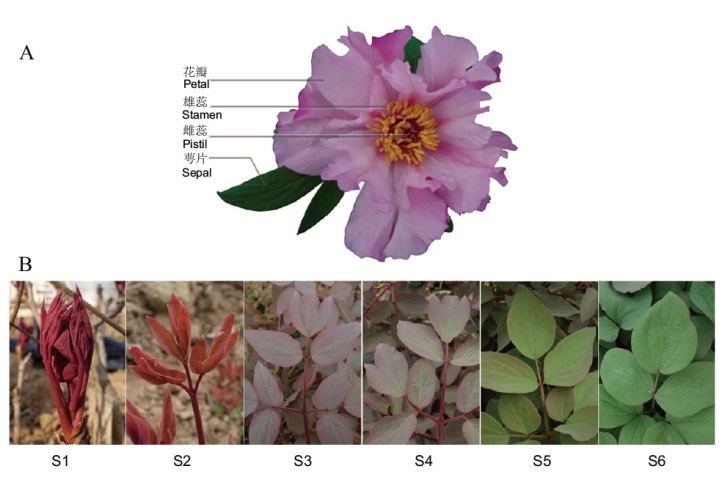
Fig. 1 Phenotypes of flowers (A) and leaves(B)of P. qiui S1: Sprout for 5 d; S2: 15 d after sprouting; S3: 35 d after sprouting; S4: 45 d after sprouting; S5: 60 d after sprouting; S6: 80 d after sprouting. The same below
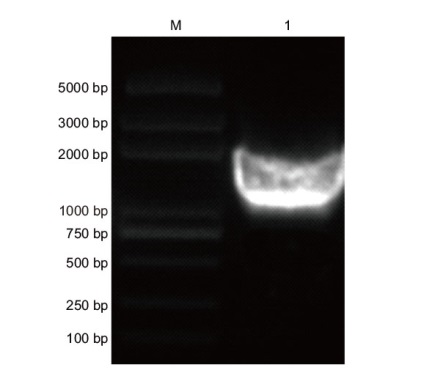
Fig. 2 PCR amplification of gene PqbHLH1 DNA fragments amplified using cDNA from peony as a template; M: DNA standards with different molecular weights; 1: amplification products of bHLH1 full-length sequence
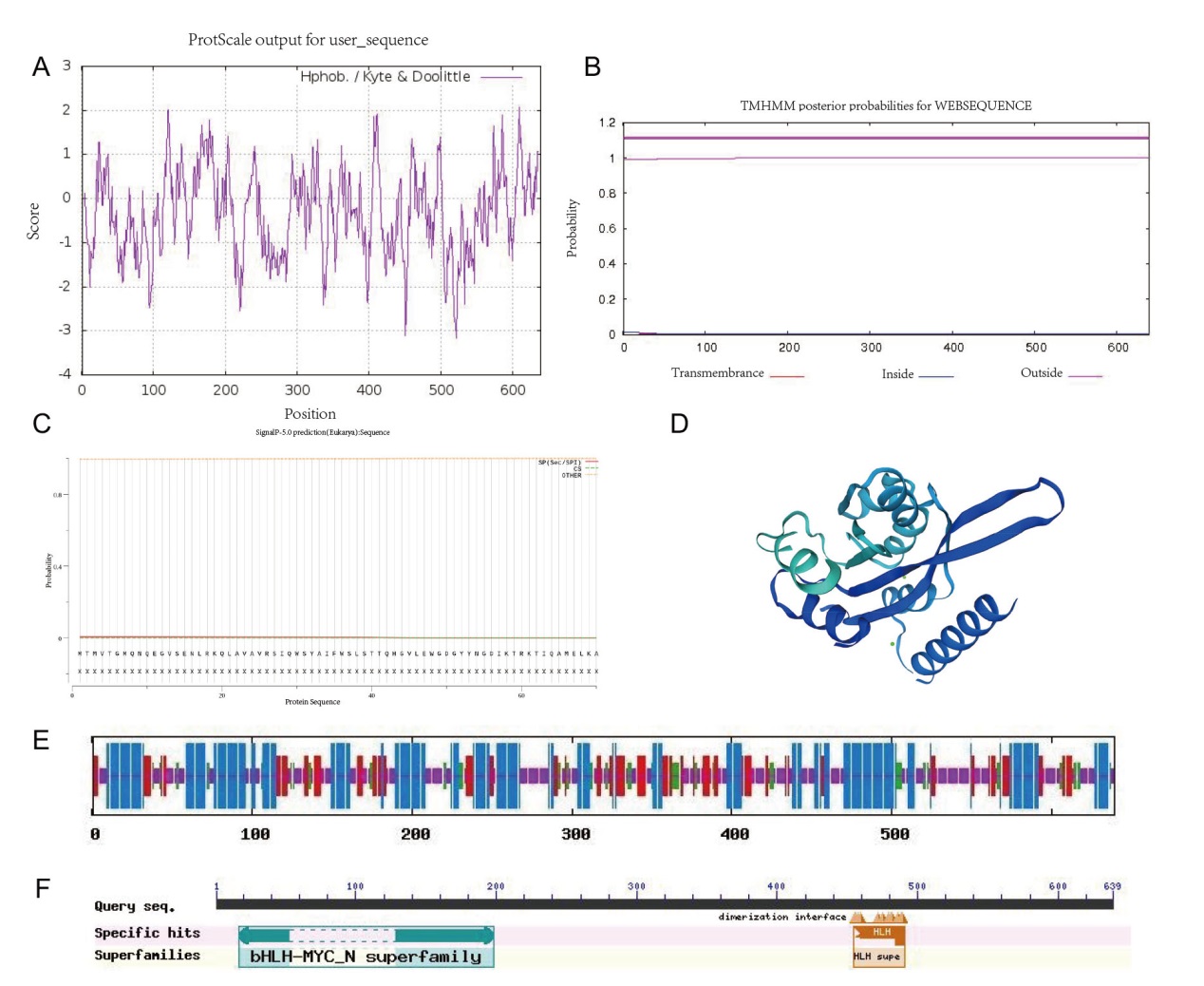
Fig. 3 Bioinformatics analysis of PqbHLH1 A: Hydrophilic and hydrophobic prediction; B: signal peptide prediction; C: amino acid transmembrane prediction; D: tertiary structure prediction of protein; E: secondary structure prediction of protein; F: structure domain analysis
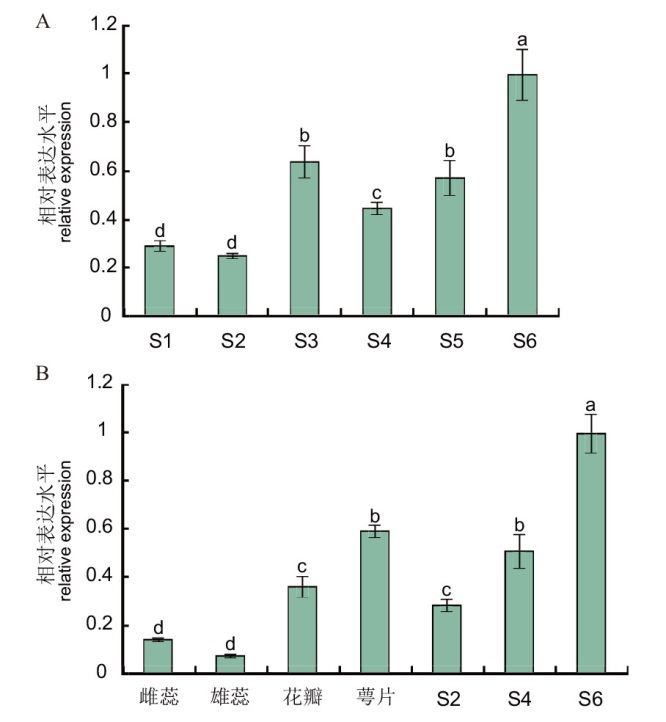
Fig. 7 Expressions of PqbHLH1 during leaf development(A)and in various tissues(B) According to the Ducan multiple test, the different letters above each column indicate significant differences, P<0.05. The same below
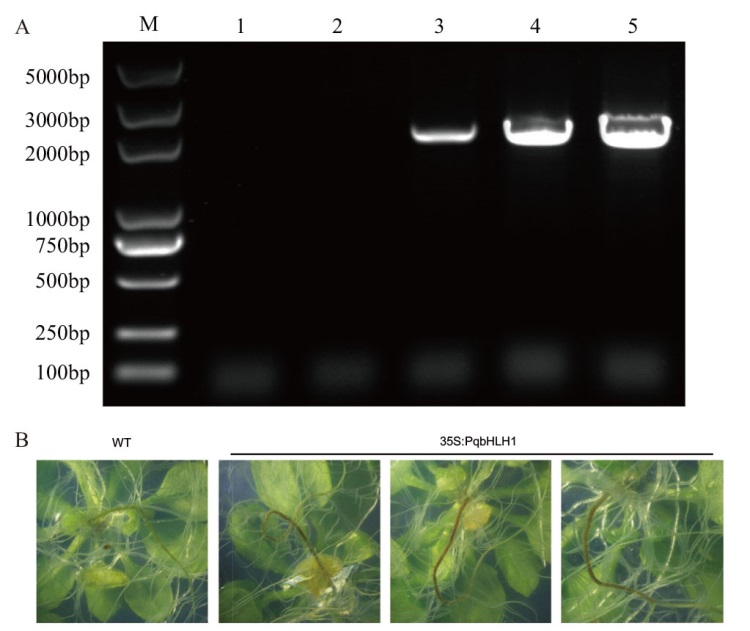
Fig. 10 PCR detection of PqbHLH1 transformed Arabi-dopsis thaliana and transgenic Arabidopsis pheno-type M:DL5000 DNA marker; 1-2: negative control; 3-5: the product PCR of transformed Arabidopsis
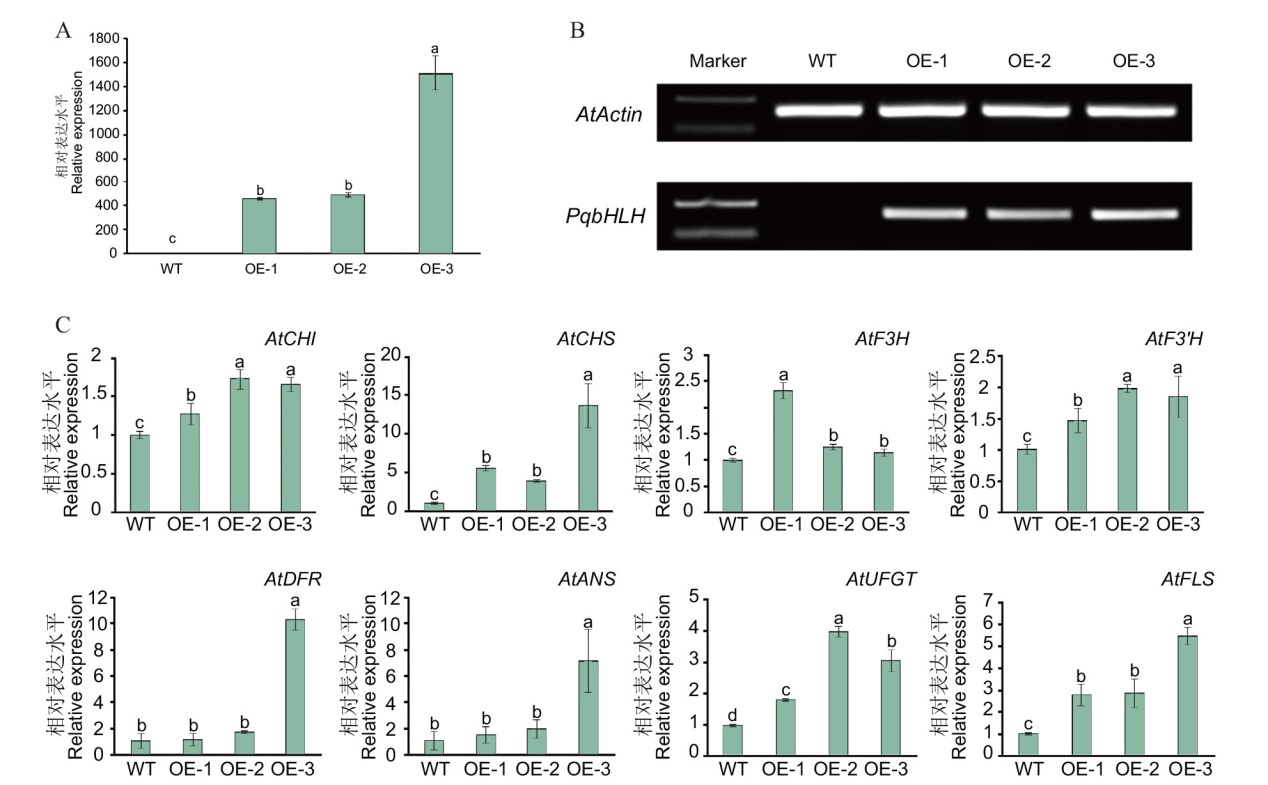
Fig. 11 Overexpression of PqbHLH1 gene promotes anthocyanin accumulation and related gene expressions in Arabidopsis A: Expression of PqbHLH1 gene in transgenic Arabidopsis; B: PCR identification of transgenic Arabidopsis(M: DNA marker; WT: wild type Arabidopsis PCR product; OE-1-3: PCR product of transgenic Arabidopsis); C: expression analysis of genes related to anthocyanin synthesis in transgenic Arabidopsis lines
| [1] | 段晶晶. 卵叶牡丹叶片花青素合成相关MYB筛选、克隆与功能验证[D]. 杨凌: 西北农林科技大学, 2019. |
| Duan JJ. Screening, cloning and functional verification of MYB related to leaf anthocyanin synthesis in Paeonia qiui[D]. Yangling: Northwest A & F University, 2019. | |
| [2] | 贾赵东, 马佩勇, 边小峰, 等. 植物花青素合成代谢途径及其分子调控[J]. 西北植物学报, 2014, 34(7): 1496-1506. |
| Jia ZD, Ma PY, Bian XF, et al. Biosynthesis metabolic pathway and molecular regulation of plants anthocyanin[J]. Acta Bot Boreali Occidentalia Sin, 2014, 34(7): 1496-1506. | |
| [3] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [4] |
高国应, 伍小方, 张大为, 等. MBW复合体在植物花青素合成途径中的研究进展[J]. 生物技术通报, 2020, 36(1): 126-134.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0738 |
| Gao GY, Wu XF, Zhang DW, et al. Research progress on the MBW complexes in plant anthocyanin biosynthesis pathway[J]. Biotechnol Bull, 2020, 36(1): 126-134. | |
| [5] | 高文, 闫庚洋, 左俊凯, 等. 蜀葵ArbHLH148基因克隆与表达分析[J]. 河南农业科学, 2023, 52(9): 133-140. |
| Gao W, Yan GY, Zuo JK, et al. Cloning and expression analysis of ArbHLH148 gene in Alcea rosea L[J]. J Henan Agric Sci, 2023, 52(9): 133-140. | |
| [6] |
Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants[J]. Mol Biol Evol, 2010, 27(4): 862-874.
doi: 10.1093/molbev/msp288 pmid: 19942615 |
| [7] | 杜杨梅. 紫花椰菜MYB-TT8-TTG1转录因子调控烟草花青素积累的研究[D]. 重庆: 西南大学, 2017. |
| Du YM. Study anthocyanin accumulation in tobacco by constitutively expression MYB2, TT8, TTG1 transcription factors from purple cauliflower[D]. Chongqing: Southwest University, 2017. | |
| [8] | 王勇江, 陈克平, 姚勤. bHLH转录因子家族研究进展[J]. 遗传, 2008, 30(7): 821-830. |
| Wang YJ, Chen KP, Yao Q. Progress of studies on bHLH transcription factor families[J]. Hereditas, 2008, 30(7): 821-830. | |
| [9] | Wu YY, Wu SH, Wang XQ, et al. Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mume[J]. Hortic Plant J, 2022, 8(4): 531-544. |
| [10] | 韩婷, 杜方. 植物萜烯类合成的转录调控研究进展[J]. 山西农业科学, 2020, 48(10): 1686-1692. |
| Han T, Du F. Research progress on regulation of transcription factors related to plant terpene sythesis[J]. J Shanxi Agric Sci, 2020, 48(10): 1686-1692. | |
| [11] | 张鹏飞, 丁楠, 李萍. 花色素苷合成酶的生物信息学分析[J]. 生物化工, 2017, 3(5): 10-15. |
| Zhang PF, Ding N, Li P. Bioinformatics analysis of anthocyanin synthesis in plants[J]. Biol Chem Eng, 2017, 3(5): 10-15. | |
| [12] |
Chandler VL, Radicella JP, Robbins TP, et al. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences[J]. Plant Cell, 1989, 1(12): 1175-1183.
pmid: 2535537 |
| [13] |
Quattrocchio F, Wing JF, van der Woude K, et al. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes[J]. Plant J, 1998, 13(4): 475-488.
doi: 10.1046/j.1365-313x.1998.00046.x pmid: 9680994 |
| [14] |
Spelt C, Quattrocchio F, Mol JN, et al. anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes[J]. Plant Cell, 2000, 12(9): 1619-1632.
doi: 10.1105/tpc.12.9.1619 pmid: 11006336 |
| [15] | Feyissa DN, Løvdal T, Olsen KM, et al. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves[J]. Planta, 2009, 230(4): 747-754. |
| [16] |
Espley RV, Hellens RP, Putterill J, et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10[J]. Plant J, 2007, 49(3): 414-427.
doi: 10.1111/j.1365-313X.2006.02964.x pmid: 17181777 |
| [17] |
Hichri I, Heppel SC, Pillet J, et al. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine[J]. Mol Plant, 2010, 3(3): 509-523.
doi: 10.1093/mp/ssp118 pmid: 20118183 |
| [18] |
Zhao PC, Li XX, Jia JT, et al. bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy[J]. J Exp Bot, 2019, 70(1): 269-284.
doi: 10.1093/jxb/ery335 pmid: 30239820 |
| [19] | Wang LH, Tang W, Hu YW, et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang[J]. Plant J, 2019, 99(2): 359-378. |
| [20] |
Su WQ, Tao R, Liu WY, et al. Characterization of four polymorphic genes controlling red leaf colour in lettuce that have undergone disruptive selection since domestication[J]. Plant Biotechnol J, 2020, 18(2): 479-490.
doi: 10.1111/pbi.13213 pmid: 31325407 |
| [21] | Wang MJ, Ou Y, Li Z, et al. Genome-wide identification and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Cymbidium ensifolium[J]. Int J Mol Sci, 2023, 24(4): 3825. |
| [22] | Deng J, Li JJ, Su MY, et al. A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis[J]. Plant Physiol Biochem, 2021, 158: 518-523. |
| [23] | Zhao R, Song XX, Yang N, et al. Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox(L.)in transgenic model plants inhibits anthocyanin accumulation[J]. Plant Cell Rep, 2020, 39(7): 891-907. |
| [24] | 宁改星. 苹果MdbHLH51和MdbHLH155的基因克隆及其在花青素合成中的作用[D]. 兰州: 甘肃农业大学, 2020. |
| Ning GX. Cloning and elucidation of the functional role of apple MdbHLH51 and MdbHLH155 in anthocyanin biosynthesis[D]. Lanzhou: Gansu Agricultural University, 2020. | |
| [25] | Park KI, Ishikawa N, Morita Y, et al. A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation[J]. Plant J, 2007, 49(4): 641-654. |
| [26] | Qi Y, Zhou L, Han LL, et al. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony(Paeonia suffruticosa)[J]. Plant Physiol Biochem, 2020, 154: 396-408. |
| [27] |
Luan YT, Tang YH, Wang X, et al. Tree peony R2R3-MYB transcription factor PsMYB30 promotes petal blotch formation by activating the transcription of the anthocyanin synthase gene[J]. Plant Cell Physiol, 2022, 63(8): 1101-1116.
doi: 10.1093/pcp/pcac085 pmid: 35713501 |
| [28] | Zhang C, Wang WN, Wang YJ, et al. Anthocyanin biosynthesis and accumulation in developing flowers of tree peony(Paeonia suffruticosa)‘Luoyang Hong’[J]. Postharvest Biol Technol, 2014, 97: 11-22. |
| [29] | 吴楠. 卵叶牡丹叶片花色苷合成相关bHLH筛选、克隆与功能分析[D]. 杨凌: 西北农林科技大学, 2021. |
| Wu N. Screening, cloning and functional analysis of bHLH related to anthocyanin synthesis of leaf in Paeonia qiui[D]. Yangling: Northwest A & F University, 2021. | |
| [30] | 朱璐璐, 周波. bHLH蛋白在植物发育及非生物胁迫中的调控[J]. 分子植物育种, 2022, 20(20): 6750-6760. |
| Zhu LL, Zhou B. Regulation of bHLH protein in plant development and abiotic stress[J]. Mol Plant Breed, 2022, 20(20): 6750-6760. | |
| [31] |
刘晓月, 王文生, 傅彬英. 植物bHLH转录因子家族的功能研究进展[J]. 生物技术进展, 2011, 1(6): 391-397.
doi: 10.3969/j.issn.2095-2341.2011.06.02 |
| Liu XY, Wang WS, Fu BY. Research progress of plant bHLH transcription factor family[J]. Curr Biotechnol, 2011, 1(6): 391-397. | |
| [32] | Hong GJ, Xue XY, Mao YB, et al. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression[J]. Plant Cell, 2012, 24(6): 2635-2648. |
| [33] | Liang YF, Ma F, Li BY, et al. A bHLH transcription factor, SlbHLH96, promotes drought tolerance in tomato[J]. Hortic Res, 2022, 9: uhac198. |
| [34] | Wang S, Zhang Z, Li LX, et al. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33[J]. Plant J, 2022, 110(4): 1021-1034. |
| [35] |
Matus JT, Poupin MJ, Cañón P, et al. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine(Vitis vinifera L.)[J]. Plant Mol Biol, 2010, 72(6): 607-620.
doi: 10.1007/s11103-010-9597-4 pmid: 20112051 |
| [36] | Lai B, Du LN, Liu R, et al. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation[J]. Front Plant Sci, 2016, 7: 166. |
| [37] | 安红立, 何霞红, 芮蕊. 林下三七PnbHLH23基因的克隆及表达模式分析[J/OL]. 分子植物育种, 2023. http://kns.cnki.net/kcms/detail/46.1068.S.20231221.1821.010.html. |
| An HL, He XH, Rui R. Cloning and expression pattern analysis of PnbHLH23 gene in Panax notoginseng under forest[J/OL]. Mol Plant Breed, 2023. http://kns.cnki.net/kcms/detail/46.1068.S.20231221.1821.010.html. | |
| [38] | Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J, 2011, 65(6): 907-921. |
| [39] | Liu ZJ, Wu XQ, Wang EH, et al. PHR1 positively regulates phosphate starvation-induced anthocyanin accumulation through direct upregulation of genes F3'H and LDOX in Arabidopsis[J]. Planta, 2022, 256(2): 42. |
| [40] | Zhu BJ, Wang Q, Wang JH, et al. DFR and PAL gene transcription and their correlation with anthocyanin accumulation in Rhodomyrtus tomentosa(Aiton.)Hassk[J]. Turk J Biochem, 2019, 44(3): 289-298. |
| [41] | Li P, Chen X, Sun F, et al. Tobacco TTG2 and ARF8 function concomitantly to control flower colouring by regulating anthocyanin synthesis genes[J]. Plant Biol, 2017, 19(4): 525-532. |
| [42] | Sun W, Shen H, Xu H, et al. Chalcone isomerase a key enzyme for anthocyanin biosynthesis in Ophiorrhiza japonica[J]. Front Plant Sci, 2019, 10: 865. |
| [43] | Li G, Michaelis DF, Huang JJ, et al. New insights into the genetic manipulation of the R2R3-MYB and CHI gene families on anthocyanin pigmentation in Petunia hybrida[J]. Plant Physiol Biochem, 2023, 203: 108000. |
| [44] | Mo RL, Han GM, Zhu ZX, et al. The ethylene response factor ERF5 regulates anthocyanin biosynthesis in ‘zijin’ mulberry fruits by interacting with MYBA and F3H genes[J]. Int J Mol Sci, 2022, 23(14): 7615. |
| [45] | Dai MJ, Kang XR, Wang YQ, et al. Functional characterization of flavanone 3-hydroxylase(F3H)and its role in anthocyanin and flavonoid biosynthesis in mulberry[J]. Molecules, 2022, 27(10): 3341. |
| [46] | 白蓓蓓, 郑燕萍, 陈黎明, 等. 不同着色火龙果成熟过程中花青素代谢途径相关酶活分析[J/OL]. 分子植物育种, 2024. http://kns.cnki.net/kcms/detail/46.1068.S.20240222.1143.004.html. |
| Bai BB, Zheng YP, Chen LM, et al. Analysis of enzyme activity related to anthocyanin metabolism pathways during the ripening process of different colored dragon fruits[J/OL]. Mol Plant Breed, 2024. http://kns.cnki.net/kcms/detail/46.1068.S.20240222.1143.004.html. | |
| [47] | Liu HL, Su BB, Zhang H, et al. Identification and functional analysis of a flavonol synthase gene from grape hyacinth[J]. Molecules, 2019, 24(8): 1579. |
| [1] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [2] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [3] | NIE Zhu-xin, GUO Jin, QIAO Zi-yang, LI Wei-wei, ZHANG Xue-yan, LIU Chun-yang, WANG Jing. Transcriptome Analysis of the Anthocyanin Biosynthesis in the Fruit Development Processes of Lycium ruthenicum Murr. [J]. Biotechnology Bulletin, 2024, 40(8): 106-117. |
| [4] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [5] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [6] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [7] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [8] | WANG Qing, NI Er-dong, WANG Qiu-shuang, QIN Dan-dan, FANG Kai-xing, LI Hong-jian, JIANG Xiao-hui, LI Bo, PAN Chen-dong, WU Hua-ling. Research Progress of UDP-glycosyltransferase Related to Anthocyanin Modification in Plants [J]. Biotechnology Bulletin, 2024, 40(7): 28-42. |
| [9] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [10] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [11] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [12] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [13] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [14] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [15] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||