








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 259-272.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0116
Previous Articles Next Articles
SHEN Zhen-hui1,2,3,4( ), CAO Yao1,2,3,4, YANG Lin-lei1,2,3,4, LUO Xiang-ying1,2,3,4, ZI Ling-shan1,2, LU Qing-qing1,2, LI Rong-chun1,2,3,4(
), CAO Yao1,2,3,4, YANG Lin-lei1,2,3,4, LUO Xiang-ying1,2,3,4, ZI Ling-shan1,2, LU Qing-qing1,2, LI Rong-chun1,2,3,4( )
)
Received:2024-01-31
Online:2024-07-26
Published:2024-07-30
Contact:
LI Rong-chun
E-mail:1947843434@qq.com;rongchunli@126.com
SHEN Zhen-hui, CAO Yao, YANG Lin-lei, LUO Xiang-ying, ZI Ling-shan, LU Qing-qing, LI Rong-chun. Cloning and Bioinformatics Analysis of the Ergothioneine Biosynthesis Genes in Naematelia aurantialba and Stereum hirsutum[J]. Biotechnology Bulletin, 2024, 40(7): 259-272.
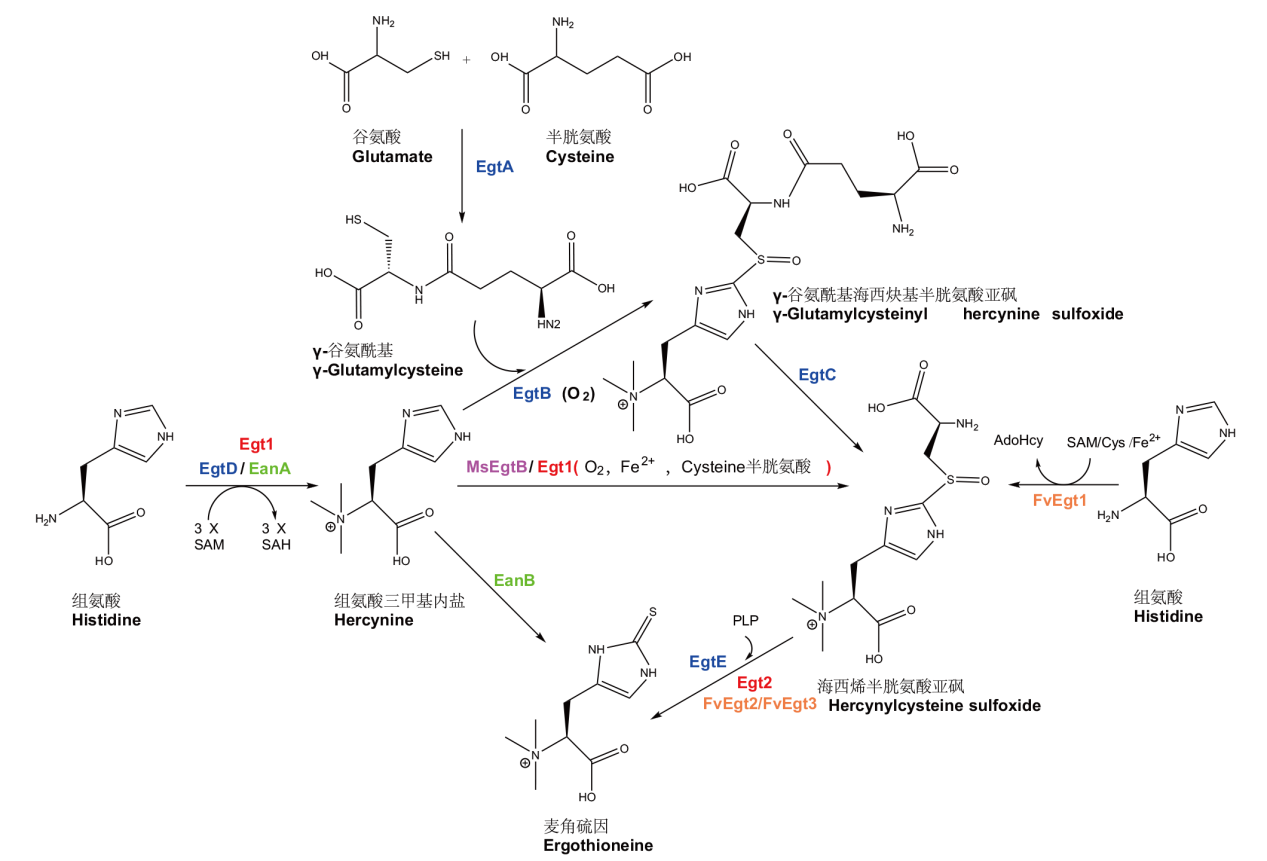
Fig. 1 Biosynthetic pathways of ergothioneine in microorganism SAM, SAH (or AdoHcy), Cys and PLP refers to s-adenosylmethionine, S-adenosylhomocysteine, cysteine and pyridoxal phosphate respectively
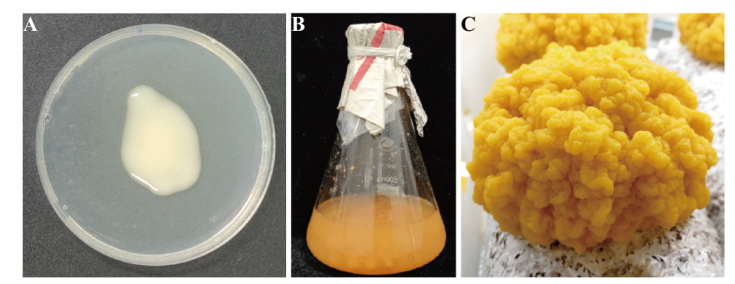
Fig. 2 Morphological characteristics of different samples A: N. aurantialba blastospore (JEYB);B: S. hirsutum liquid culture(ShFJY+ShJST);C: N. aurantialba fruiting body(JEZST)
| 序号Serial No. | 引物名称 Primer name | 序列 Sequence(5'-3') | 退火温度 Annealing temperature/℃ |
|---|---|---|---|
| 1 | ShEgt1-F | TGCGTCGGTCTCCTTTCTT | 53 |
| 2 | ShEgt1-R | CGGAGCAGAACAGATTTATCG | |
| 3 | ShEgt2-F | CACCACCACACATCACAGTC | 56 |
| 4 | ShEgt2-R | GACAAGTTCACAACGAGCAT | |
| 5 | NaEgt1-F | TGTCTGTCTCCTCCATCCA | 56 |
| 6 | NaEgt1-R | TAGTCCCAGGCTCATCACA | |
| 7 | NaEgt2-F | GTATCGCTGTTTACCACCTGT | 56 |
| 8 | NaEgt2-F | TCTACTCATCCTTCGTCGG |
Table 1 Primer information for ergothioneine biosynthetic genes of N. aurantialba and S. hirsutum
| 序号Serial No. | 引物名称 Primer name | 序列 Sequence(5'-3') | 退火温度 Annealing temperature/℃ |
|---|---|---|---|
| 1 | ShEgt1-F | TGCGTCGGTCTCCTTTCTT | 53 |
| 2 | ShEgt1-R | CGGAGCAGAACAGATTTATCG | |
| 3 | ShEgt2-F | CACCACCACACATCACAGTC | 56 |
| 4 | ShEgt2-R | GACAAGTTCACAACGAGCAT | |
| 5 | NaEgt1-F | TGTCTGTCTCCTCCATCCA | 56 |
| 6 | NaEgt1-R | TAGTCCCAGGCTCATCACA | |
| 7 | NaEgt2-F | GTATCGCTGTTTACCACCTGT | 56 |
| 8 | NaEgt2-F | TCTACTCATCCTTCGTCGG |
| 序号Serial No. | 分析软件Analysis software | 用途Application | 网址Web site |
|---|---|---|---|
| 1 | ORF | CDS序列预测CDS sequence prediction | |
| 2 | Gene Structure Display Server | 绘制基因结构图Drawing a genetic structure map | |
| 3 | NCBI-Blast | 基因同源性分析Gene homology analysis | |
| 4 | ExPASy- ProtParam | 分析蛋白的理化性质Analyzing the physicochemical properties of proteins | |
| 5 | SignalP4.1 | 信号肽预测Prediction of signal peptide | |
| 6 | PSORT II | 亚细胞定位Subcellular localization | |
| 7 | CCD | 保守结构域预测Conservative structural domain prediction | |
| 8 | PSIPRED | 二级结构预测Secondary structure of protein prediction | |
| 9 | SWISS-MODEL | 三级结构预测Tertiary structure prediction | |
| 10 | Phyre2 | 三级结构配体预测Tertiary structure ligand prediction | |
| 11 | MEGA7.0 | 系统进化树分析Phylogenetic analysis | |
Table 2 Bioinformatics software used in this study
| 序号Serial No. | 分析软件Analysis software | 用途Application | 网址Web site |
|---|---|---|---|
| 1 | ORF | CDS序列预测CDS sequence prediction | |
| 2 | Gene Structure Display Server | 绘制基因结构图Drawing a genetic structure map | |
| 3 | NCBI-Blast | 基因同源性分析Gene homology analysis | |
| 4 | ExPASy- ProtParam | 分析蛋白的理化性质Analyzing the physicochemical properties of proteins | |
| 5 | SignalP4.1 | 信号肽预测Prediction of signal peptide | |
| 6 | PSORT II | 亚细胞定位Subcellular localization | |
| 7 | CCD | 保守结构域预测Conservative structural domain prediction | |
| 8 | PSIPRED | 二级结构预测Secondary structure of protein prediction | |
| 9 | SWISS-MODEL | 三级结构预测Tertiary structure prediction | |
| 10 | Phyre2 | 三级结构配体预测Tertiary structure ligand prediction | |
| 11 | MEGA7.0 | 系统进化树分析Phylogenetic analysis | |
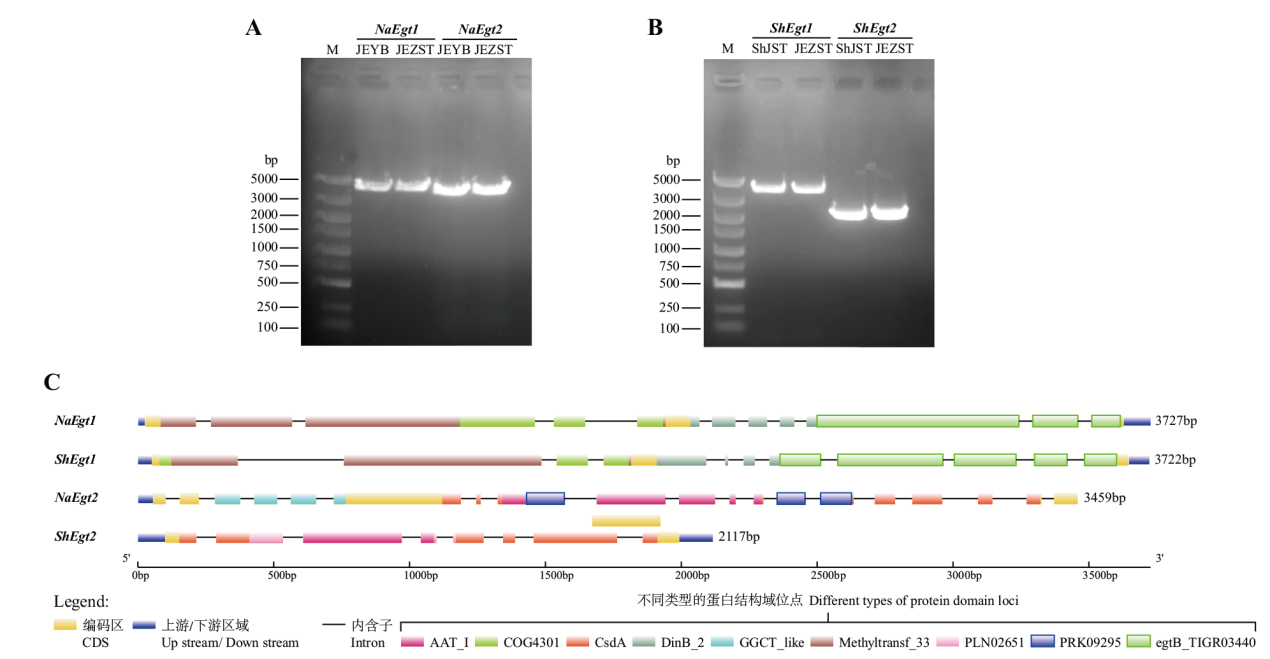
Fig. 3 Verification of PCR amplification of Egt1 and Egt2 genes of N. aurantialba and S. hirsutum(A, B)and gene structure analysis(C) M: DL2000 marker; ShJST: S. hirsutum mycelium; JEYB: N. aurantialba blastospore ; JEZST: N. aurantialba fruiting body
| 基因 Gene | CDS长度CDS length/bp | 蛋白质长度 Protein length | 蛋白质分子量Molecular weight/kD | 等电点 Isoelectric point | 总平均亲水系数Grand average of hydropathicity (GRAVY) | 信号肽 Signal peptide | 细胞中分布 Location |
|---|---|---|---|---|---|---|---|
| NaEgt1 | 2952 | 984 | 110.32 | 5.30 | -0.479 | 无No | 细胞质Cytoplasmic |
| ShEgt1 | 2565 | 885 | 99.03 | 5.19 | -0.528 | 无No | 细胞质Cytoplasmic |
| NaEgt2 | 2178 | 726 | 81.61 | 5.87 | -0.368 | 无No | 细胞质Cytoplasmic |
| ShEgt2 | 1383 | 461 | 51.46 | 5.68 | -0.238 | 无No | 细胞质Cytoplasmic |
Table 3 Analysis of physical and chemical properties of Egt1 and Egt2 in two species
| 基因 Gene | CDS长度CDS length/bp | 蛋白质长度 Protein length | 蛋白质分子量Molecular weight/kD | 等电点 Isoelectric point | 总平均亲水系数Grand average of hydropathicity (GRAVY) | 信号肽 Signal peptide | 细胞中分布 Location |
|---|---|---|---|---|---|---|---|
| NaEgt1 | 2952 | 984 | 110.32 | 5.30 | -0.479 | 无No | 细胞质Cytoplasmic |
| ShEgt1 | 2565 | 885 | 99.03 | 5.19 | -0.528 | 无No | 细胞质Cytoplasmic |
| NaEgt2 | 2178 | 726 | 81.61 | 5.87 | -0.368 | 无No | 细胞质Cytoplasmic |
| ShEgt2 | 1383 | 461 | 51.46 | 5.68 | -0.238 | 无No | 细胞质Cytoplasmic |

Fig. 4 Tertiary structure and binding site analysis of five fungal Egt1 and Egt2 proteins A: Comparison of the tertiary structures of five fungal Egt1 and Egt2(Egt3); B: Egt1 binds to imidazole(red arrow)and acetate ions(blue arrow); C: binding of Egt1 to s-adenosylmethionine(SAM); D: the combination of EgtD and Fe3+; E: Egt1 binds to cysteine(red arrow); F: Egt2 binds to S-mercaptocysteine; G-H: Egt2 binds to two pyridoxal phosphate(PLP)sites. SpEgt1, NcEgt1, FvEgt1, SpEgt2, NcEgt2, FvEgt2 and FvEgt3 are key enzyme for ergothioneine biosynthesis of S. pombe, N. crassa and F. filiformis, respectively(Accession number: CAA22334.2, XP_956324.3, QBB19872.1, NP_595091.1, A7UX13, QBB19873.1 and QBB19874.1).
| 蛋白 Protein | 物种 Species | 模板 Template | 覆盖度 Coverage/% | 可信度 Confidence/% | 功能域一致性 Functional domain identity/% | 结合位点 Binding site |
|---|---|---|---|---|---|---|
| Egt1 | 耻垢分枝杆菌 M smegmatis | c4uy5A | 51/46/55/36/73 | 100/100/100/100/100 | 34/32/26/24/35 | 咪唑和醋酸根离子 Imidazole and acetate ion |
| 大利什曼原虫 Leishmania major | d1xtpa | 23/35/38/-/59 | 96/98/99/-/98 | 31/30/18/-/19 | s-腺苷甲硫氨酸 S-adenosyl-methionine(SAM) | |
| 耐热分枝杆菌 Mycobacterium thermoresistibile | c4x8bA | 44/49/55/57/- | 100/100/100/100/- | 27/26/23/24/- | Fe3+离子 Fe3+ ion | |
| 嗜热氢单胞菌 Hydrogenimonas thermophila | c8khqD | 44/48/55/57/- | 100/100/100/100/- | 26/26/24/24/- | 半胱氨酸 Cysteine(Cys) | |
| Egt2 | 粗糙脉孢霉 N. crassa | c5utsC | 59/91/97/94/93/92 | 100/100/100/100/100/100 | 39/35/31/97/21/36 | - |
| 集胞藻属 Synechocystis sp. | d1elua | 56/88/95/90/90/89 | 99.9/100/100/100/100/100 | 20/19/16/16/18/16 | 磷酸吡哆醛和S-巯基半胱氨酸 Pyridoxal phosphate and S-mercaptocysteine |
Table 4 Predicted information on the tertiary structure of Egt1 and Egt2 proteins from five fungal species
| 蛋白 Protein | 物种 Species | 模板 Template | 覆盖度 Coverage/% | 可信度 Confidence/% | 功能域一致性 Functional domain identity/% | 结合位点 Binding site |
|---|---|---|---|---|---|---|
| Egt1 | 耻垢分枝杆菌 M smegmatis | c4uy5A | 51/46/55/36/73 | 100/100/100/100/100 | 34/32/26/24/35 | 咪唑和醋酸根离子 Imidazole and acetate ion |
| 大利什曼原虫 Leishmania major | d1xtpa | 23/35/38/-/59 | 96/98/99/-/98 | 31/30/18/-/19 | s-腺苷甲硫氨酸 S-adenosyl-methionine(SAM) | |
| 耐热分枝杆菌 Mycobacterium thermoresistibile | c4x8bA | 44/49/55/57/- | 100/100/100/100/- | 27/26/23/24/- | Fe3+离子 Fe3+ ion | |
| 嗜热氢单胞菌 Hydrogenimonas thermophila | c8khqD | 44/48/55/57/- | 100/100/100/100/- | 26/26/24/24/- | 半胱氨酸 Cysteine(Cys) | |
| Egt2 | 粗糙脉孢霉 N. crassa | c5utsC | 59/91/97/94/93/92 | 100/100/100/100/100/100 | 39/35/31/97/21/36 | - |
| 集胞藻属 Synechocystis sp. | d1elua | 56/88/95/90/90/89 | 99.9/100/100/100/100/100 | 20/19/16/16/18/16 | 磷酸吡哆醛和S-巯基半胱氨酸 Pyridoxal phosphate and S-mercaptocysteine |

Fig. 5 Phylogenetic tree analysis of Egt1(A)and Egt2(B)in different species Purple colours indicate the results of Egt2 sequence comparison between S. hirsutum and N. aurantialba; red colours indicate the results of Egt2 sequence comparison between S. hirsutum and other fungi; blue colours indicate the results of Egt2 sequence comparison between N. aurantialba and other fungi
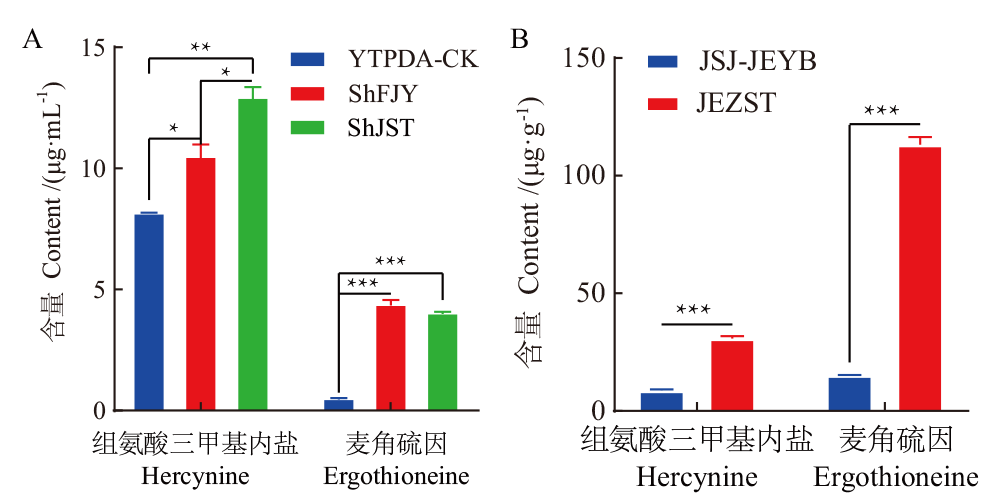
Fig. 7 Determination of hercynine and ergothioneine content in different samples A: Liquid sample; B: solid sample. YTPDA-CK: PDA liquid medium(control); ShFJY: S. hirsutum fermentation broth; ShJST: S. hirsutum mycelium; JSJ-JEYB: N. aurantialba blastospa; JEZST: N. aurantialba fruiting body. *, ** and *** indicate a significant difference at P<0.05, P<0.01 and P<0.0001, respectively
| [1] | Tanret C. Surune base nouvelle retiree du seigle ergote, L-ergoth-ioneine compt[J]. Rend Acad Sci, 1909, 149: 222-224. |
| [2] |
Franzoni F, Colognato R, Galetta F, et al. An in vitro study on the free radical scavenging capacity of ergothioneine: comparison with reduced glutathione, uric acid and trolox[J]. Biomed Pharmacother, 2006, 60(8): 453-457.
pmid: 16930933 |
| [3] | 林陈水, 付水星, 黎小军, 等. 一种稀有的天然氨基酸-麦角硫因[J]. 氨基酸和生物资源, 2006, 28(1): 63-67. |
| Lin CS, Fu SX, Li XJ, et al. Ergothioneine—a rare natural amino acid[J]. Amino Acids Biotic Resour, 2006, 28(1): 63-67. | |
| [4] | Speisky H, Gómez M, Carrasco-Pozo C, et al. Cu(I)-glutathione complex: a potential source of superoxide radicals generation[J]. Bioorg Med Chem, 2008, 16(13): 6568-6574. |
| [5] | 朱本占, 毛莉, 范瑞梅, 等. 天然抗氧化剂麦角硫因保护铜所致DNA和蛋白质氧化损伤的作用机理[J]. 科学通报, 2011, 56(27): 2283-2288. |
| Zhu BZ, Mao L, Fan RM, et al. Mechanism of protection by natural antioxidant ergothioneine against copper-induced oxidative damage to DNA and protein[J]. Chin Sci Bull, 2011, 56(27): 2283-2288. | |
| [6] | Markova NG, Karaman-Jurukovska N, Dong KK, et al. Skin cells and tissue are capable of using L-ergothioneine as an integral component of their antioxidant defense system[J]. Free Radic Biol Med, 2009, 46(8): 1168-1176. |
| [7] | 张萌萌, 张倩, 张国琛. 金针菇提取液中麦角硫因氨基酸的功能及其在食品工业中的应用前景[J]. 安徽农业科学, 2014, 42(11): 3385-3387, 3390. |
| Zhang MM, Zhang Q, Zhang GC. The function of ergothioneine in the extracts from Flammulina velutipes and its application prospects on food industry[J]. J Anhui Agric Sci, 2014, 42(11): 3385-3387, 3390. | |
| [8] | 张翠, 赵艳敏, 白淑芳, 等. HPLC法测定不同品种蘑菇中麦角硫因的含量[J]. 食品工业科技, 2013, 34(23): 307-310. |
| Zhang C, Zhao YM, Bai SF, et al. HPLC determination of ergothioneine in mushrooms of different species[J]. Sci Technol Food Ind, 2013, 34(23): 307-310. | |
| [9] | 木开代斯·买合木提, 陈建, 焦春伟, 等. L-麦角硫因生物合成与应用研究进展[J]. 天然产物研究与开发, 2022, 34(4): 713-721. |
| Mukaidaisi·MHMT, Chen J, Jiao CW, et al. Progress in biosynthesis and application of L-ergothioneine[J]. Nat Prod Res Dev, 2022, 34(4): 713-721. | |
| [10] | 刘琦, 毛雨丰, 廖小平, 等. 麦角硫因生物合成研究的新进展[J]. 生物工程学报, 2022, 38(4): 1408-1420. |
| Liu Q, Mao YF, Liao XP, et al. Recent progress in ergothioneine biosynthesis: a review[J]. Chin J Biotechnol, 2022, 38(4): 1408-1420. | |
| [11] | Seebeck FP. In vitro reconstitution of mycobacterial ergothioneine biosynthesis[J]. J Am Chem Soc, 2010, 132(19): 6632-6633. |
| [12] | Kamide T, Takusagawa S, Tanaka N, et al. High Production of Ergothioneine in Escherichia coli using the Sulfoxide Synthase from Methylobacterium strains[J]. J Agric Food Chem, 2020, 68(23): 6390-6394. |
| [13] | Burn R, Misson L, Meury M, et al. Anaerobic origin of ergothioneine[J]. Angew Chem Int Ed Engl, 2017, 56(41): 12508-12511. |
| [14] | Bello MH, Barrera-Perez V, Morin D, et al. The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination[J]. Fungal Genet Biol, 2012, 49(2): 160-172. |
| [15] | Pluskal T, Ueno M, Yanagida M. Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system[J]. PLoS One, 2014, 9(5): e97774. |
| [16] |
Fujitani Y, Alamgir KM, Tani A. Ergothioneine production using Methylobacterium species, yeast, and fungi[J]. J Biosci Bioeng, 2018, 126(6): 715-722.
doi: S1389-1723(18)30295-0 pmid: 29910189 |
| [17] | Hu W, Song H, Sae Her A, et al. Bioinformatic and biochemical characterizations of C-S bond formation and cleavage enzymes in the fungus Neurospora crassa ergothioneine biosynthetic pathway[J]. Org Lett, 2014, 16(20): 5382-5385. |
| [18] | Cumming BM, Chinta KC, Reddy VP, et al. Role of ergothioneine in microbial physiology and pathogenesis[J]. Antioxid Redox Signal, 2018, 28(6): 431-444. |
| [19] | Yang XQ, Lin SX, Lin JD, et al. The biosynthetic pathway of ergothioneine in culinary-medicinal winter mushroom, Flammulina velutipes(agaricomycetes)[J]. Int J Med Mushrooms, 2020, 22(2): 171-181. |
| [20] | 梅保良, 刘琦, 姜文侠, 等. 营养因子强化麦角硫因生物合成的研究[J]. 食品研究与开发, 2015, 36(15): 108-112. |
| Mei BL, Liu Q, Jiang WX, et al. Study on the biosynthesis of L-ergothioneine by enhancement of nutritional factors[J]. Food Res Dev, 2015, 36(15): 108-112. | |
| [21] | Tepwong P, Giri A, Sasaki F, et al. Mycobial enhancement of ergothioneine by submerged cultivation of edible mushroom mycelia and its application as an antioxidative compound[J]. Food Chem, 2012, 131(1): 247-258. |
| [22] | Dubost NJ, Beelman RB, Royse DJ. Influence of selected cultural factors and postharvest storage on ergothioneine content of common button mushroom Agaricus bisporus(J. lge)imbach(agaricomycetideae)[J]. Int J Med Mushr, 2007, 9(2): 163-176. |
| [23] | Liang CH, Huang LY, Ho KJ, et al. Submerged cultivation of mycelium with high ergothioneine content from the culinary-medicinal king oyster mushroom Pleurotus eryngii(higher basidiomycetes)and its composition[J]. Int J Med Mushrooms, 2013, 15(2): 153-164. |
| [24] |
Kalaras MD, Richie JP, Calcagnotto A, et al. Mushrooms: a rich source of the antioxidants ergothioneine and glutathione[J]. Food Chem, 2017, 233: 429-433.
doi: S0308-8146(17)30691-X pmid: 28530594 |
| [25] | Lin SY, Chien SC, Wang SY, et al. Submerged cultivation of My-celium with high ergothioneine content from the culinary-medicinal golden oyster mushroom, Pleurotus citrinopileatus(higher basidiomycetes)[J]. Int J Med Mushrooms, 2015, 17(8): 749-761. |
| [26] | 杨林雷, 李荣春, 曹瑶, 等. 金耳的学名及分类地位考证[J]. 食药用菌, 2020, 28(4): 252-255, 276. |
| Yang LL, Li RC, Cao Y, et al. Research on the scientific Name and taxonomic status of “Jin Er”[J]. Edible Med Mushrooms, 2020, 28(4): 252-255, 276. | |
| [27] |
Liu XZ, Wang QM, Göker M, et al. Towards an integrated phylogenetic classification of the Tremellomycetes[J]. Stud Mycol, 2015, 81: 85-147.
doi: 10.1016/j.simyco.2015.12.001 pmid: 26955199 |
| [28] | 郭正堂. 中国韧革菌(Ⅲ)[J]. 植物研究, 1987, 7(3): 85-112. |
| Guo ZT. Stereaceae in China(Ⅲ)[J]. Bull Bot Res, 1987, 7(3): 85-112. | |
| [29] | 罗晓莉, 张沙沙, 曹晶晶, 等. 云南3种胶质食用菌营养成分分析与蛋白质营养价值评价[J]. 食品工业科技, 2021, 42(14): 328-333. |
| Luo XL, Zhang SS, Cao JJ, et al. Analysis of nutritional components and evaluation of protein nutritional value of three kinds of gelatinous edible fungi in Yunnan[J]. Sci Technol Food Ind, 2021, 42(14): 328-333. | |
| [30] | 曹瑶, 李荣春, 杨林雷, 等. 工厂化栽培金耳的氨基酸组成及蛋白质营养评价[J]. 食药用菌, 2021, 29(2): 152-156. |
| Cao Y, Li RC, Yang LL, et al. Amino acid composition and nutritional evaluation of protein of industrial cultivated Naematelia aurantialba[J]. Edible Med Mushrooms, 2021, 29(2): 152-156. | |
| [31] | 曹瑶, 李荣春, 杨林雷, 等. 工厂化栽培金耳的营养成分测定及品质评价[J]. 食药用菌, 2021, 29(4): 318-322. |
| Cao Y, Li RC, Yang LL, et al. Basic nutrition component analysis and quality evaluation of industrial cultivated “Jiner”(Naematelia aurantialba)[J]. Edible Med Mushrooms, 2021, 29(4): 318-322. | |
| [32] | 曹瑶, 杨晓君, 杨林雷, 等. 金耳浆对挂面品质的影响[J]. 食品工程, 2023(3): 27-30, 34. |
| Cao Y, Yang XJ, Yang LL, et al. Effects of the fresh mushroom of N. aurantialba on the noodle quality[J]. Food Eng, 2023(3): 27-30, 34. | |
| [33] | 沈真辉, 罗祥英, 赵仙伟, 等. 利用转录组分析光照对金耳子实体转色的影响[J]. 食用菌学报, 2023, 30(6): 12-27. |
| Shen ZH, Luo XY, Zhao XW, et al. Transcriptome analysis revealed impact of light quality on coloration of Naematelia aurantialba fruiting bodies[J]. Acta Edulis Fungi, 2023, 30(6): 12-27. | |
| [34] | 杨林雷, 李荣春, 曹瑶, 等. 金耳及金耳多糖的药用保健功效及其机理研究进展[J]. 食药用菌, 2021, 29(3): 176-182. |
| Yang LL, Li RC, Cao Y, et al. Research progress on the pharmacology and health care effects of Naematelia aurantialba and its polysaccharides[J]. Edible Med Mushrooms, 2021, 29(3): 176-182. | |
| [35] | 何容, 罗晓莉, 李建英, 等. 金耳研究现状与展望[J]. 食药用菌, 2019, 27(1): 41-47. |
| He R, Luo XL, Li JY, et al. Research status and prospect of Tremella aurantialba[J]. Edible Med Mushrooms, 2019, 27(1): 41-47. | |
| [36] | 孙涛, 姜浩, 王燕玲, 等. 金耳多糖的研究进展[J]. 中国食品学报, 2022, 22(8): 386-397. |
| Sun T, Jiang H, Wang YL, et al. Research advances on Naematelia aurantialba polysaccharides[J]. J Chin Inst Food Sci Technol, 2022, 22(8): 386-397. | |
| [37] | Yan YH, Wang MT, Chen N, et al. Isolation, structures, bioactivities, application and future prospective for polysaccharides from Tremella aurantialba: a review[J]. Front Immunol, 2022, 13: 1091210. |
| [38] |
Halliwell B, Cheah IK, Tang RMY. Ergothioneine - a diet-derived antioxidant with therapeutic potential[J]. FEBS Lett, 2018, 592(20): 3357-3366.
doi: 10.1002/1873-3468.13123 pmid: 29851075 |
| [39] | 薛天凯, 赵艳敏, 林纪伟, 等. 正交设计优化平菇下脚料中麦角硫因的提取工艺[J]. 食品研究与开发, 2017, 38(3): 40-45. |
| Xue TK, Zhao YM, Lin JW, et al. Optimization of the extraction technique of ergothioneine from mushroom scraps by orthogonal method[J]. Food Res Dev, 2017, 38(3): 40-45. | |
| [40] | 赵艳敏, 雷智东, 刘成航, 等. 高效液相色谱法测定多种真菌中麦角硫因含量[J]. 食品研究与开发, 2016, 37(2): 117-119. |
| Zhao YM, Lei ZD, Liu CH, et al. Determination of ergothioneine in multiple species of fungi by HPLC[J]. Food Res Dev, 2016, 37(2): 117-119. | |
| [41] | 陈柏雄. 蛹虫草虫草素和麦角硫因生物合成基因功能及其调控机制研究[D]. 广州: 华南农业大学, 2020. |
| Chen BX. The biosynthetic genes fuction and regulatory mechanism of cordycepin and ergothioneine in Cordyceps militaris[D]. Guangzhou: South China Agricultural University, 2020. | |
| [42] | 林金德. 侧耳属食用菌麦角硫因合成酶基因的挖掘及其功能研究[D]. 广州: 华南农业大学, 2020. |
| Lin JD. Mining and function study of ergothioneine synthase genes of Pleurotus edible fungus[D]. Guangzhou: South China Agricultural University, 2020. | |
| [43] | 余颖豪. 灰树花麦角硫因生物合成基因的功能研究[D]. 广州: 华南农业大学, 2020. |
| Yu YH. Elucidation of the function of ergothioneine biosynthesis genes from Grifola frondosa[D]. Guangzhou: South China Agricultural University, 2020. | |
| [44] | Jeong JH, Cha HJ, Ha SC, et al. Structural insights into the histidine trimethylation activity of EgtD from Mycobacterium smegmatis[J]. Biochem Biophys Res Commun, 2014, 452(4): 1098-1103. |
| [1] | PANG Meng-zhen, XU Han-qin, LIU Hai-yan, SONG Juan, WANG Jia-han, SUN Li-na, JI Pei-mei, YIN Ze-zhi, HU You-chuan, ZHAO Xiao-meng, LIANG Shan-shan, ZHANG Si-ju, LUAN Wei-jiang. Gene Identification and Functional Analysis of Yellowish and Early Heading Mutant hz1 in Rice [J]. Biotechnology Bulletin, 2024, 40(7): 125-136. |
| [2] | HUANG Dan, JIANG Shan, PENG Tao. Cloning of FfCYP98 Gene and Its Functional Analysis in Folioceros fuciformis [J]. Biotechnology Bulletin, 2024, 40(7): 273-284. |
| [3] | HE Yu-bing, FU Zhen-hao, LI Ren-han, LIU Xiu-xia, LIU Chun-li, YANG Yan-kun, LI Ye, BAI Zhong-hu. Efficient Biosynthesis of 2-Naphthaleneethanol in Metabolically Engineered Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2024, 40(7): 99-107. |
| [4] | HU Jin-jin, LI Su-zhen, MA Xu-hui, LIU Xiao-qing, XIE Shan-shan, JIANG Hai-yang, CHEN Ru-mei. Regulation of Maize Anthocyanin Biosynthesis Metabolism [J]. Biotechnology Bulletin, 2024, 40(6): 34-44. |
| [5] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [6] | HAO Si-yi, ZHANG Jun-ke, WANG Bin, QU Peng-yan, LI Rui-de, CHENG Chun-zhen. Cloning and Expression Analysis of Banana EARLY FLOWERING 3(ELF3)Genes [J]. Biotechnology Bulletin, 2024, 40(5): 131-140. |
| [7] | DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis [J]. Biotechnology Bulletin, 2024, 40(4): 122-129. |
| [8] | LIU Huan-huan, YANG Li-chun, LI Huo-gen. Cloning and Functional Analysis of LtMYB305 in Liriodendron tulipifera [J]. Biotechnology Bulletin, 2024, 40(4): 179-188. |
| [9] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [10] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [11] | WANG Jun-fang, HUANG Qiu-bin, ZHANG Piao-dan, ZHANG Peng-pai. Structure and Biosynthesis of Surfactin as well as Its Role in Biological Control [J]. Biotechnology Bulletin, 2024, 40(1): 100-112. |
| [12] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [13] | CHEN Zhi-min, LI Cui, WEI Ji-tian, LI Xin-ran, LIU Yi, GUO Qiang. Research Progress in the Regulation of Chlorogenic Acid Biosynthesis and Its Application [J]. Biotechnology Bulletin, 2024, 40(1): 57-71. |
| [14] | LI Liang, XU Shan-shan, JIANG Yan-jun. Research Progress in the Production of Ergothioneine by Biosynthesis [J]. Biotechnology Bulletin, 2024, 40(1): 86-99. |
| [15] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||