








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 273-284.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0068
Previous Articles Next Articles
HUANG Dan1( ), JIANG Shan2(
), JIANG Shan2( ), PENG Tao1(
), PENG Tao1( )
)
Received:2024-01-16
Online:2024-07-26
Published:2024-07-30
Contact:
JIANG Shan, PENG Tao
E-mail:1093617061@qq.com;kyosan200312@hotmail.com;pengtao@gznu.edu.cn
HUANG Dan, JIANG Shan, PENG Tao. Cloning of FfCYP98 Gene and Its Functional Analysis in Folioceros fuciformis[J]. Biotechnology Bulletin, 2024, 40(7): 273-284.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| JB-F | ATGAGGAGTTTGCGAAGCA |
| JB-R | CCTTCCCCTCGTCGTCCAC |
| 3'RACE 特异性引物F1 | AGGGGCCGCAGTCTCATTTCGT |
| 3'RACE 特异性引物F2 | CTCCTGGGGCTGCAGAAGCAGT |
| 5'RACE 特异性引物R1 | AACGCGAGGCGCGTAATGTTGT |
| 5'RACE 特异性引物R2 | GCCGAGAGGTACGTCTTGGGGTT |
| FfCYP98-F | TCTAGAATGGGAGCAACTCTGAACGT |
| FfCYP98-R | GGATCCGACGGTTTTGGCATACAAGG |
| HACtin-F | ACGCCTGAACATCTCCTGAA |
| HACtin-R | CGTGCAGAACAAGAACTCGT |
| qpcr-F | CGCAGTCTCATTTCGTGGAC |
| qpcr-R | TCCTTCTGCACCTTCCTCTG |
| 跨载体-F | ccaaccacgtcttcaaagca |
| 跨载体-R | atccagactgaatgcccaca |
| ACtion2-F | GGAAGGATCTGTACGGTAAC |
| ACtion2-R | GGACCTGCCTCATCTTACT |
| Action-F | TTCAATGTCCCTGCCATGTATG |
| Action-R | AATACCGGTTGTACGACCAC |
| PAL-F | TCTGCGAAAAGGATTTACTCAAAGTT |
| PAL-R | CTACAAGGATCATCCGCGTATGT |
| C4H-F | CCTCCAGGACAGTCTAAAGTGGATACTA |
| C4H-R | CGATTATGGAGTGGTTAAGGATGTG |
| HCT-F | TCTTTATGGGACCTGGTGGAATT |
| HCT-R | GATAAGCTGCCATCATTAGTAGGACTTG |
| 4CL-F | TCAACCCGGTGAGATTTGTA |
| 4CL-R | TCGTCATCGATCAATCCAAT |
| C3H-F | AAGTCTAGTGGAGCGAAACAGCATT |
| C3H-R | TCCTCACTAAGATCATACTGATCCTTCA |
| CAD-F | TGCGATTGGATCCGATGGAA |
| CAD-R | GCTTCCCTGCCTCAGTCATA |
Table 1 Primer sequences used in the study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| JB-F | ATGAGGAGTTTGCGAAGCA |
| JB-R | CCTTCCCCTCGTCGTCCAC |
| 3'RACE 特异性引物F1 | AGGGGCCGCAGTCTCATTTCGT |
| 3'RACE 特异性引物F2 | CTCCTGGGGCTGCAGAAGCAGT |
| 5'RACE 特异性引物R1 | AACGCGAGGCGCGTAATGTTGT |
| 5'RACE 特异性引物R2 | GCCGAGAGGTACGTCTTGGGGTT |
| FfCYP98-F | TCTAGAATGGGAGCAACTCTGAACGT |
| FfCYP98-R | GGATCCGACGGTTTTGGCATACAAGG |
| HACtin-F | ACGCCTGAACATCTCCTGAA |
| HACtin-R | CGTGCAGAACAAGAACTCGT |
| qpcr-F | CGCAGTCTCATTTCGTGGAC |
| qpcr-R | TCCTTCTGCACCTTCCTCTG |
| 跨载体-F | ccaaccacgtcttcaaagca |
| 跨载体-R | atccagactgaatgcccaca |
| ACtion2-F | GGAAGGATCTGTACGGTAAC |
| ACtion2-R | GGACCTGCCTCATCTTACT |
| Action-F | TTCAATGTCCCTGCCATGTATG |
| Action-R | AATACCGGTTGTACGACCAC |
| PAL-F | TCTGCGAAAAGGATTTACTCAAAGTT |
| PAL-R | CTACAAGGATCATCCGCGTATGT |
| C4H-F | CCTCCAGGACAGTCTAAAGTGGATACTA |
| C4H-R | CGATTATGGAGTGGTTAAGGATGTG |
| HCT-F | TCTTTATGGGACCTGGTGGAATT |
| HCT-R | GATAAGCTGCCATCATTAGTAGGACTTG |
| 4CL-F | TCAACCCGGTGAGATTTGTA |
| 4CL-R | TCGTCATCGATCAATCCAAT |
| C3H-F | AAGTCTAGTGGAGCGAAACAGCATT |
| C3H-R | TCCTCACTAAGATCATACTGATCCTTCA |
| CAD-F | TGCGATTGGATCCGATGGAA |
| CAD-R | GCTTCCCTGCCTCAGTCATA |

Fig. 2 Bioinformatics analysis of the FfCYP98 A: Physicochemical characterization and secondary structure prediction of FfCYP98. B: Tertiary structure prediction of FfCYP98. C: Analysis of hydrophilicity and hydrophobicity of FfCYP98 amino acid sequences. D: Analysis of the transmembrane structural domains of FfCYP98. E: Phylogenetic tree of FfCYP98

Fig. 4 Identification of transgenic A. thaliana and relative expressions of total phenols, total flavonoids and phenylpropane synthesis pathway genes in transgenic A. thaliana A: Resistance screening for Arabidopsis thaliana. B: Identification of transgenic A. thaliana, M: Marker = 15 000 bp, 1: Control, 2: WT. C: Expression of the FfCYP98 gene intransgenic A. thaliana(***P< 0.001,****P< 0.0001). D: Detection of total phenols and total flavonoids in transgenic A. thaliana. E: Expression analysis of transgenic A. thaliana phenylpropane synthesis pathway genes
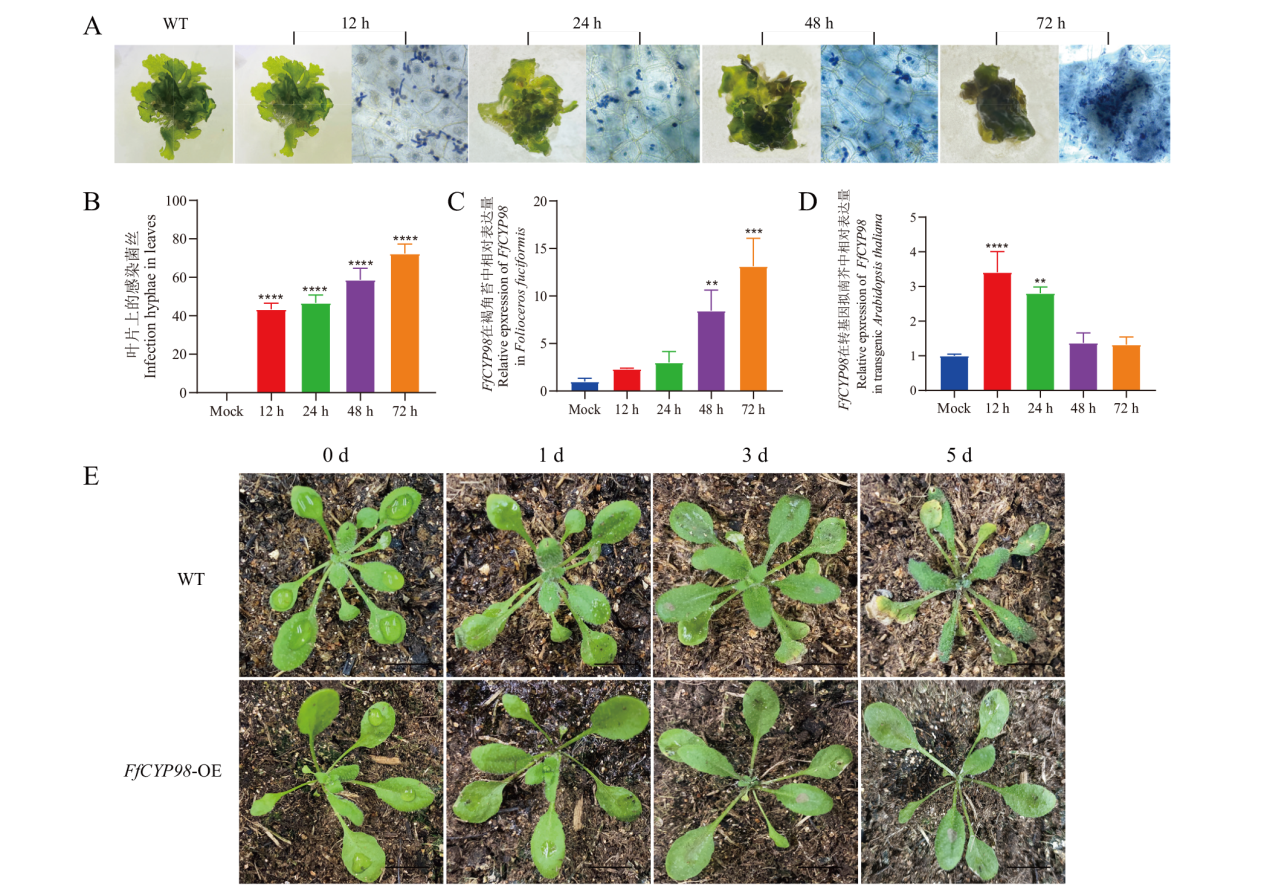
Fig. 5 Infected behavior of B. cinerea on F. fuciformis and characterization of FfCYP98 expression in transgenic A. thaliana under B. cinerea infestation A: B. cinerea infests Folioceros fuciformis, bar=100 μm. B: Comparison of the invasion rate of hyphae in F. fuciformis. C: Expression of FfCYP98 at different times of F. fuciformis under the stress of B. cinerea. D: Observation of transgenic A. thaliana infection with B. cinerea, bar= 1 cm. E: Expressions of FfCYP98 at different times of transgenic A. thaliana under the stress of B. cinerea, **P< 0.01, ***P< 0.001, ****P< 0.0001
| [1] | Kenrick P, Wellman CH, Schneider H, et al. A timeline for terrestrialization: consequences for the carbon cycle in the Palaeozoic[J]. Philos Trans R Soc Lond B Biol Sci, 2012, 367(1588): 519-536. |
| [2] |
Yin RH, Ulm R. How plants cope with UV-B: from perception to response[J]. Curr Opin Plant Biol, 2017, 37: 42-48.
doi: S1369-5266(16)30228-X pmid: 28411583 |
| [3] | Vanholme R, Storme V, Vanholme B, et al. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis[J]. Plant Cell, 2012, 24(9): 3506-3529. |
| [4] |
Davies KM, Jibran R, Zhou YF, et al. The evolution of flavonoid biosynthesis: a bryophyte perspective[J]. Front Plant Sci, 2020, 11: 7.
doi: 10.3389/fpls.2020.00007 pmid: 32117358 |
| [5] | Alber AV, Renault H, Basilio-Lopes A, et al. Evolution of coumaroyl conjugate 3-hydroxylases in land plants: lignin biosynthesis and defense[J]. Plant J, 2019, 99(5): 924-936. |
| [6] | Kono Y, Kashine S, Yoneyama T, et al. Iron chelation by chlorogenic acid as a natural antioxidant[J]. Biosci Biotechnol Biochem, 1998, 62(1): 22-27. |
| [7] | Clé C, Hill LM, Niggeweg R, et al. Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance[J]. Phytochemistry, 2008, 69(11): 2149-2156. |
| [8] |
Barbehenn R, Dukatz C, Holt C, et al. Feeding on poplar leaves by caterpillars potentiates foliar peroxidase action in their guts and increases plant resistance[J]. Oecologia, 2010, 164(4): 993-1004.
doi: 10.1007/s00442-010-1733-y pmid: 20680646 |
| [9] |
Escamilla-Treviño LL, Shen H, Hernandez T, et al. Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass(Panicum virgatum L.)[J]. Plant Mol Biol, 2014, 84(4-5): 565-576.
doi: 10.1007/s11103-013-0152-y pmid: 24190737 |
| [10] |
Vanholme R, Cesarino I, Rataj K, et al. Caffeoyl shikimate esterase(CSE)is an enzyme in the lignin biosynthetic pathway in Arabidopsis[J]. Science, 2013, 341(6150): 1103-1106.
doi: 10.1126/science.1241602 pmid: 23950498 |
| [11] | 王雪霞, 薛永常, 赵文超. 木质素生物合成中C3H/HCT的研究进展[J]. 生命的化学, 2008, 28(5): 650-653. |
| Wang XX, Xue YC, Zhao WC. The research progress of C3H/HCT in lignin biosynthesis[J]. Chem Life, 2008, 28(5): 650-653. | |
| [12] |
Renault H, De Marothy M, Jonasson G, et al. Gene duplication leads to altered membrane topology of a cytochrome P450 enzyme in seed plants[J]. Mol Biol Evol, 2017, 34(8): 2041-2056.
doi: 10.1093/molbev/msx160 pmid: 28505373 |
| [13] | 胡人亮. 漫话苔藓[J]. 植物杂志, 1978(4): 41-43. |
| Hu RL. Manga Moss[J]. Plants, 1978(4): 41-43. | |
| [14] |
Basile A, Giordano S, López-Sáez JA, et al. Antibacterial activity of pure flavonoids isolated from mosses[J]. Phytochemistry, 1999, 52(8): 1479-1482.
pmid: 10647220 |
| [15] | Umezawa T. Diversity in lignan biosynthesis[J]. Phytochem Rev, 2003, 2(3): 371-390. |
| [16] | Xu ZY, Zhang DD, Hu J, et al. Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom[J]. BMC Bioinformatics, 2009, 10(Suppl 11): S3. |
| [17] | Jiang S, Tian X, Huang XL, et al. Physcomitrium patens CAD1 has distinct roles in growth and resistance to biotic stress[J]. BMC Plant Biol, 2022, 22(1): 518. |
| [18] | Garcia C, Sérgio C, Villarreal JC, et al. The HornwortsDendrocerosNees andMegacerosCampb. in São tomé e Príncipe(africa, gulf of Guinea)with the description of Dendroceros paivaesp. nov[J]. Cryptogam Bryol, 2012, 33(1): 3-21. |
| [19] | Renzaglia KS, Villarreal JC, Duff RJ, et al. New insights into morphology, anatomy, and systematics of hornworts[M]//Shaw AJ, ed. Bryophyte Biology. Cambridge: Cambridge University Press, 2010: 139-172. |
| [20] | Qiu YL, Li LB, Wang B, et al. The deepest divergences in land plants inferred from phylogenomic evidence[J]. Proc Natl Acad Sci U S A, 2006, 103(42): 15511-15516. |
| [21] | Ernst L, Wohl J, Bauerbach E, et al. Hydroxycinnamoyltransferase and CYP98 in phenolic metabolism in the rosmarinic acid-producing hornwort Anthoceros agrestis[J]. Planta, 2022, 255(4): 75. |
| [22] |
Zhang J, Fu XX, Li RQ, et al. The hornwort genome and early land plant evolution[J]. Nat Plants, 2020, 6(2): 107-118.
doi: 10.1038/s41477-019-0588-4 pmid: 32042158 |
| [23] |
刘阳星月, 张迪, 姚亚亚, 等. 大豆过敏原11S球蛋白G2中A2链结合表位的预测[J]. 食品科学, 2018, 39(18): 152-158.
doi: 10.7506/spkx1002-6630-201818024 |
| Liu YXY, Zhang D, Yao YY, et al. Mapping of ig G-binding epitopes on the A2 chain of the major soybean allergen 11S globulin G2[J]. Food Sci, 2018, 39(18): 152-158. | |
| [24] | 何姗. 农杆菌介导CmWRKY15-1基因对菊花的遗传转化[D]. 沈阳: 沈阳农业大学, 2020. |
| He S. Agrobacterium-mediated genetic transformation of CmWRKY15-1 gene to Chrysanthemum[D]. Shenyang: Shenyang Agricultural University, 2020. | |
| [25] | 魏加练. 基于转录组的粗山羊草响应镉胁迫关键基因挖掘及AetHIPP28的功能验证[D]. 贵阳: 贵州师范大学, 2022. |
| Wei JL. Identification and the function of Aegilops tauschii AetHIPP28 to cadmium stress based on transcriptome sequencing[D]. Guiyang: Guizhou Normal University, 2022. | |
| [26] | 李保华, 赵美琦. 梨叶抗黑星病作用机制研究[J]. 莱阳农学院学报, 2003, 20(3): 187-188. |
| Li BH, Zhao MQ. Resistant mechanism of pear leaves against Venturia nashicola[J]. J Laiyang Agric Coll, 2003, 20(3): 187-188. | |
| [27] |
Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacte-rium-mediated transformation of Arabidopsis thaliana[J]. Plant J, 1998, 16(6): 735-743.
doi: 10.1046/j.1365-313x.1998.00343.x pmid: 10069079 |
| [28] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [29] | 王殿, 庄亚妹, 徐华, 等. 甘蓝型油菜CCCH基因BnaA07g26050D的克隆及表达分析[J]. 山东师范大学学报: 自然科学版, 2017, 32(4): 137-143. |
| Wang D, Zhuang YM, Xu H, et al. Cloning and expression of the ccch gene bnaa07g26050d in brassica napus L[J]. J Shandong Norm Univ Nat Sci, 2017, 32(4): 137-143. | |
| [30] |
张蕊杰, 顾茁, 薛辰旭, 等. 青稞萌芽富集酚类物质品种筛选及条件优化[J]. 食品与发酵工业, 2022, 48(15): 193-200.
doi: 10.13995/j.cnki.11-1802/ts.028750 |
| Zhang RJ, Gu Z, Xue CX, et al. Screening of highland barley varieties using phenolics accumulation as indicator and the effect of germination conditions on phenolics[J]. Food Ferment Ind, 2022, 48(15): 193-200. | |
| [31] | Dong NQ, Lin HX. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions[J]. J Integr Plant Biol, 2021, 63(1): 180-209. |
| [32] | Mizutani M, Ward E, DiMaio J, et al. Molecular cloning and sequencing of a cDNA encoding mung bean cytochrome P450(P450C4H)possessing cinnamate 4-hydroxylase activity[J]. Biochem Biophys Res Commun, 1993, 190(3): 875-880. |
| [33] |
Teutsch HG, Hasenfratz MP, Lesot A, et al. Isolation and sequence of a cDNA encoding the Jerusalem artichoke cinnamate 4-hydroxylase, a major plant cytochrome P450 involved in the general phenylpropanoid pathway[J]. Proc Natl Acad Sci USA, 1993, 90(9): 4102-4106.
doi: 10.1073/pnas.90.9.4102 pmid: 8097885 |
| [34] |
Lavhale SG, Kalunke RM, Giri AP. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants[J]. Planta, 2018, 248(5): 1063-1078.
doi: 10.1007/s00425-018-2965-z pmid: 30078075 |
| [35] | Chen XH, Wang HT, Li XY, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4(4CL-like 1)from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis[J]. BMC Plant Biol, 2019, 19(1): 231. |
| [36] | Costa MA, Collins RE, Anterola AM, et al. An in silico assessment of gene function and organization of the phenylpropanoid pathway metabolic networks in Arabidopsis thaliana and limitations thereof[J]. Phytochemistry, 2003, 64(6): 1097-1112. |
| [37] | 潘琛, 任百光, 盖颖. 对香豆酰辅酶A酯生物合成及纯化方法[J]. 北京林业大学学报, 2016, 38(3): 120-124. |
| Pan C, Ren BG, Gai Y. Method of enzymatic synthesis and purification of p-coumaroyl-CoA[J]. J Beijing For Univ, 2016, 38(3): 120-124. | |
| [38] | 李洋, 唐雪冰, 李晓峰, 等. NtC3H基因对烟草类黄酮及绿原酸合成的影响[J]. 中国烟草科学, 2016, 37(1): 8-13. |
| Li Y, Tang XB, Li XF, et al. The influence of NtC3H on the synthesis of flavonoids and chlorogenic acid in tobacco[J]. Chin Tob Sci, 2016, 37(1): 8-13. | |
| [39] | Xin JK, Che TM, Huang XL, et al. A comprehensive view of metabolic responses to CYP98 perturbation in ancestral plants[J]. Plant Physiol Biochem, 2023, 201: 107793. |
| [40] | 高梅. 小立碗藓4- 香豆酰莽草酸/喹啉3'-羟化酶基因功能的初步研究[D]. 贵阳: 贵州师范大学, 2020. |
| Gao M. A preliminary study on the gene function of 4-coumaruylshikimate/quinoline 3'-hydroxylase[D]. Guiyang: Guizhou Normal University, 2020. | |
| [41] |
Boerjan W, Ralph J, Baucher M. Lignin biosynthesis[J]. Annu Rev Plant Biol, 2003, 54: 519-546.
pmid: 14503002 |
| [42] | 何利, 唐翠明, 戴凡炜, 等. 防御酶与植物抗青枯病关系研究进展[J]. 广东农业科学, 2013, 40(12): 101-103. |
| He L, Tang CM, Dai FW, et al. Relationship between the defense enzyme and the disease resistance of plants to bacterial wilt[J]. Guangdong Agric Sci, 2013, 40(12): 101-103. | |
| [43] | Li C, Zhang MM, Wei S, et al. Study of Walnut brown rot caused by Botryosphaeria dothidea in the Guizhou Province of China[J]. Crop Prot, 2023, 163: 106118. |
| [44] | Wang D, Xu H, Huang JY, et al. The Arabidopsis CCCH protein C3H14 contributes to basal defense against Botrytis cinerea mainly through the WRKY33-dependent pathway[J]. Plant Cell Environ, 2020, 43(7): 1792-1806. |
| [1] | PANG Meng-zhen, XU Han-qin, LIU Hai-yan, SONG Juan, WANG Jia-han, SUN Li-na, JI Pei-mei, YIN Ze-zhi, HU You-chuan, ZHAO Xiao-meng, LIANG Shan-shan, ZHANG Si-ju, LUAN Wei-jiang. Gene Identification and Functional Analysis of Yellowish and Early Heading Mutant hz1 in Rice [J]. Biotechnology Bulletin, 2024, 40(7): 125-136. |
| [2] | FAN Zong-qiang, FENG Jing-han, ZHENG Li-xue, WANG Shuo, PENG Xiang-qian, CHEN Fang. Study on the Control and Induced Resistance in Cucumber with Bacillus subtilis B579 against Cucumber Fusarium Wilt [J]. Biotechnology Bulletin, 2024, 40(7): 226-234. |
| [3] | SHEN Zhen-hui, CAO Yao, YANG Lin-lei, LUO Xiang-ying, ZI Ling-shan, LU Qing-qing, LI Rong-chun. Cloning and Bioinformatics Analysis of the Ergothioneine Biosynthesis Genes in Naematelia aurantialba and Stereum hirsutum [J]. Biotechnology Bulletin, 2024, 40(7): 259-272. |
| [4] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [5] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [6] | HAO Si-yi, ZHANG Jun-ke, WANG Bin, QU Peng-yan, LI Rui-de, CHENG Chun-zhen. Cloning and Expression Analysis of Banana EARLY FLOWERING 3(ELF3)Genes [J]. Biotechnology Bulletin, 2024, 40(5): 131-140. |
| [7] | DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis [J]. Biotechnology Bulletin, 2024, 40(4): 122-129. |
| [8] | LIU Huan-huan, YANG Li-chun, LI Huo-gen. Cloning and Functional Analysis of LtMYB305 in Liriodendron tulipifera [J]. Biotechnology Bulletin, 2024, 40(4): 179-188. |
| [9] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [10] | PENG Yu-jia, LI Wen-cui, LIU Yong-bo. Research Progress in the Evolution Mechanisms for Insect Resistance to Insecticides and Bt-transgenic Plants [J]. Biotechnology Bulletin, 2024, 40(4): 40-51. |
| [11] | FANG Tian-yi, YUE Yan-ling. Mechanisms of Root Exudates-mediated Plant Resistance to Soil-borne Diseases [J]. Biotechnology Bulletin, 2024, 40(3): 52-61. |
| [12] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [13] | LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 233-244. |
| [14] | ZHU Tian-yi, KONG Gui-mei, JIAO Hong-mei, GUO Ting-ting, WU Ri-han, LIU Cui-cui, GAO Cheng-feng, LI Guo-cai. Establishment of A Bacterial Model of CRISPR/Cas9 Mediated adeG Gene Knockout in Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(2): 55-64. |
| [15] | ZOU Xiu-wei, YUE Jia-ni, LI Zhi-yu, DAI Liang-ying, LI Wei. Functional Analysis of Rice Heat Shock Transcription Factor HsfA2b Regulating the Resistance to Abiotic Stresses [J]. Biotechnology Bulletin, 2024, 40(2): 90-98. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||