








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (1): 240-251.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0469
Previous Articles Next Articles
KOU Bei-sen1,2,3( ), CHENG Meng-meng1,2,3, GUO Xue-qin1,2,3, GE Bin1,2,3, LIU Di1,2,3, LU Hai1,2,3, LI Hui1,2,3(
), CHENG Meng-meng1,2,3, GUO Xue-qin1,2,3, GE Bin1,2,3, LIU Di1,2,3, LU Hai1,2,3, LI Hui1,2,3( )
)
Received:2024-05-20
Online:2025-01-26
Published:2025-01-22
Contact:
LI Hui
E-mail:koubeisen@163.com;lihui830@bjfu.edu.cn
KOU Bei-sen, CHENG Meng-meng, GUO Xue-qin, GE Bin, LIU Di, LU Hai, LI Hui. Effects of Histone Deacetylase Inhibitor TSA Treatment on the Stem Development of Poplar[J]. Biotechnology Bulletin, 2025, 41(1): 240-251.
| 样品 Simple | 原始数据Raw reads(×106) | 高质量数据Clean reads(×106) | Q20/% | Q30/% | 比对到参考基因组 Alignment to reference genome /% | 唯一参考序列数 Number of unique reference sequences/% | 比对到参考基因组 Alignment to reference genome/% | 唯一参考序列数 Number of unique reference sequences/% |
|---|---|---|---|---|---|---|---|---|
| WT0 | 47.77 | 42.32 | 97.2 | 91.3 | 96.69 | 84.51 | 96.69 | 84.51 |
| CL2 | 48.93 | 42.84 | 97.3 | 91.8 | 96.44 | 84.38 | 96.44 | 84.38 |
| CL12 | 46.02 | 42.46 | 97.1 | 91.4 | 96.98 | 85.22 | 96.98 | 85.22 |
Table 1 Details of transcriptome sequencing data
| 样品 Simple | 原始数据Raw reads(×106) | 高质量数据Clean reads(×106) | Q20/% | Q30/% | 比对到参考基因组 Alignment to reference genome /% | 唯一参考序列数 Number of unique reference sequences/% | 比对到参考基因组 Alignment to reference genome/% | 唯一参考序列数 Number of unique reference sequences/% |
|---|---|---|---|---|---|---|---|---|
| WT0 | 47.77 | 42.32 | 97.2 | 91.3 | 96.69 | 84.51 | 96.69 | 84.51 |
| CL2 | 48.93 | 42.84 | 97.3 | 91.8 | 96.44 | 84.38 | 96.44 | 84.38 |
| CL12 | 46.02 | 42.46 | 97.1 | 91.4 | 96.98 | 85.22 | 96.98 | 85.22 |
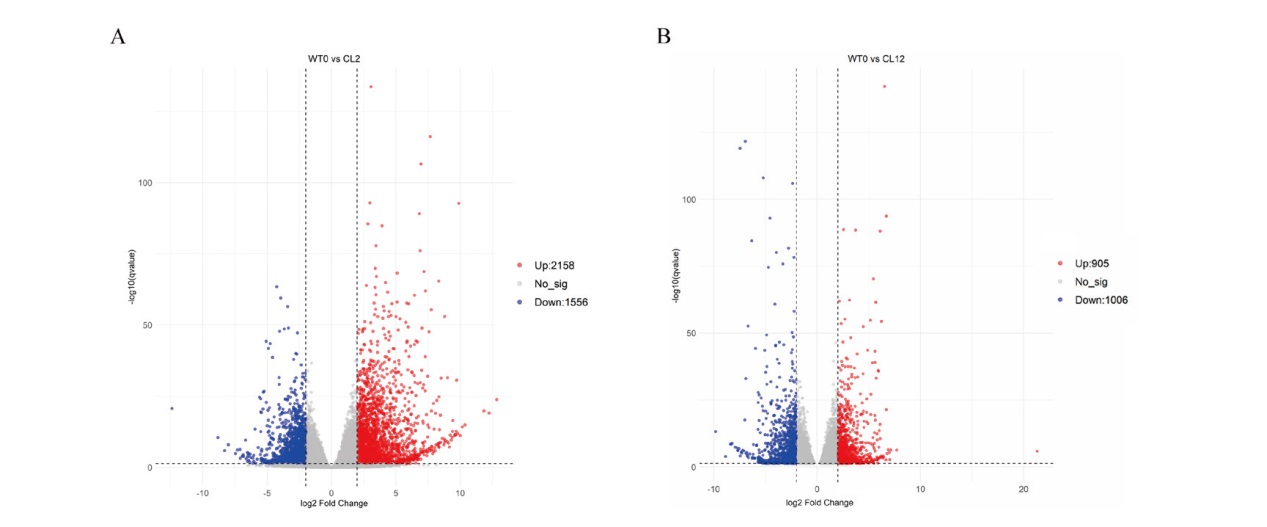
Fig. 2 Statistical data of DEGs A: Volcano plot of DEGs between WT0 and CL2. B: Volcano plot of DEGs between WT0 and CL12. Red dots: Up-regulated DEGs. Blue dots: Down-regulated DEGs. Gray dots: Non- DEGs
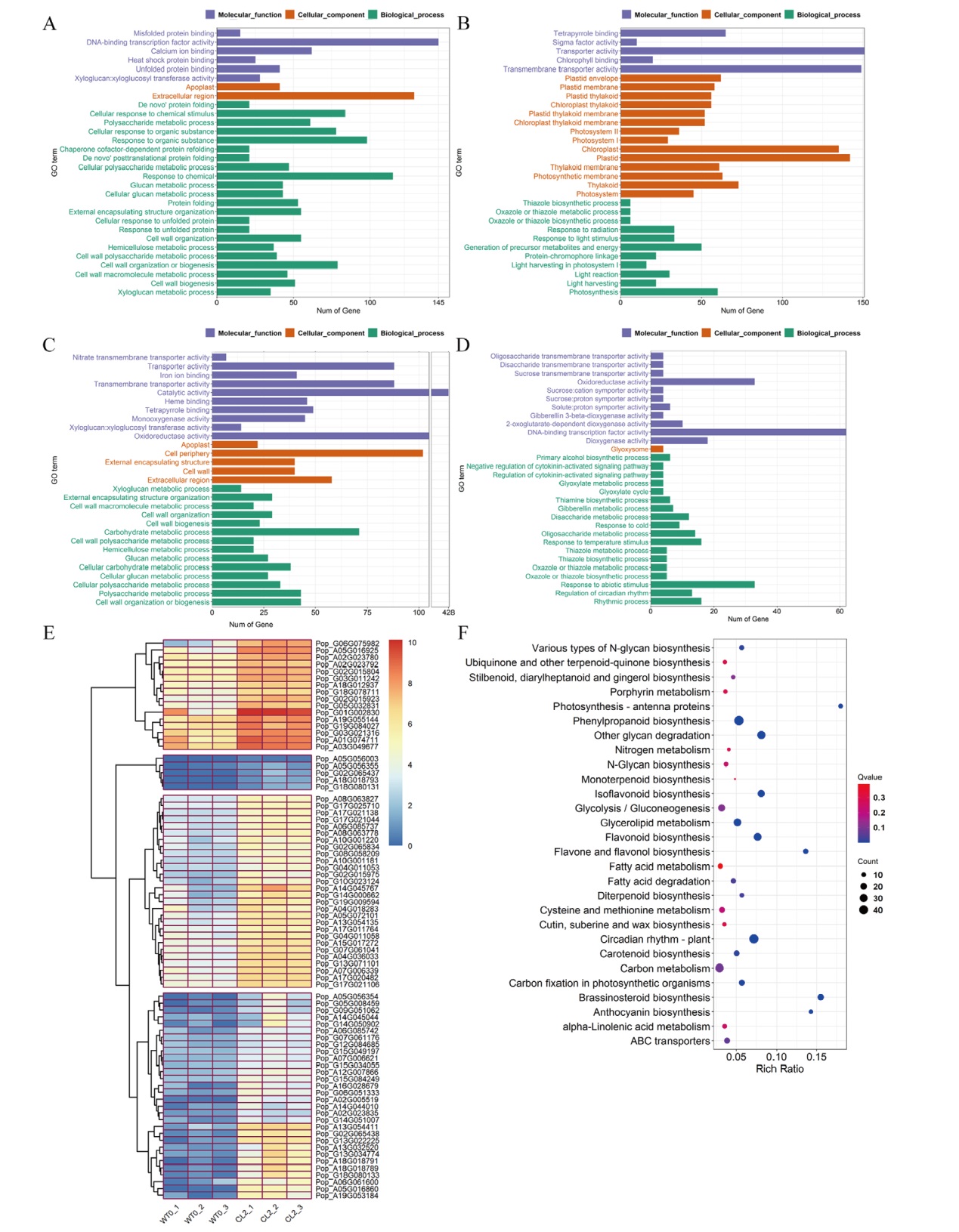
Fig. 3 GO functional enrichment histogram, KEGG pathway enrichment histogram and expression clustering heat map of DEGs A-B: GO enrichment histogram of 2 h up-regulated(A)and down-regulated(B)DEGs. C-D: GO enrichment histogram of 12 h up-regulated(C)and down-regulated(D)DEGs. E: Cluster heat map of DEGS associated with cell wall development. F: KEGG enrichment histogram of 12 h up-regulated DEGs
| 基因ID Gene ID | log2 (CL2/WT0) | Q value (WT0-vs-CL2) | 描述 Description |
|---|---|---|---|
| Pop_A01G003964 | 5.769956725 | 1.32E-41 | ERF1A |
| Pop_A01G032260 | 5.649644852 | 1.26E-05 | ERF21 |
| Pop_A01G059811 | 5.539774145 | 5.91E-07 | ERF61 |
| Pop_A01G060003 | 7.0861912 | 1.99E-09 | DREB1E |
| Pop_A04G018519 | 3.029867059 | 3.13E-06 | bHLH162 |
| Pop_A06G061940 | 2.433933971 | 0.032353534 | ORG2 |
| Pop_A14G044066 | 2.36624488 | 0.001496264 | bZip53 |
| Pop_A08G063663 | 2.588359391 | 4.82E-13 | CDF2 |
| Pop_G19G009429 | 2.356323627 | 6.09E-16 | CDF3 |
| Pop_G09G015087 | 2.033526993 | 0.003029387 | ZAT10 |
| Pop_A16G090138 | 3.342912788 | 3.50E-13 | PHL7 |
| Pop_G10G047973 | 6.429548834 | 0.001027136 | ORR25 |
| Pop_A09G014447 | 2.372666749 | 6.59E-06 | SCR3 |
| Pop_A06G062082 | 6.325653816 | 0.001158459 | HSF4b |
| Pop_A16G055308 | 2.667183627 | 5.18E-06 | HSF3 |
| Pop_A02G066179 | 5.110284639 | 8.26E-05 | MYB73 |
| Pop_A07G011878 | 2.013444428 | 1.37E-12 | MYB44 |
| Pop_A08G063813 | 2.04957107 | 0.013470953 | MYB108 |
| Pop_A14G000372 | 3.81186488 | 1.23E-08 | MYB44 |
| Pop_A15G064557 | 4.679506153 | 1.94E-16 | MYB62 |
| Pop_A04G019588 | 2.206753814 | 6.93E-06 | NAC73 |
| Pop_A04G026457 | 3.711825186 | 0.000111049 | TCP13 |
| Pop_A01G059075 | 3.408636654 | 6.29E-05 | WRKY28 |
| Pop_A01G080041 | 3.413796772 | 6.86E-30 | WRKY40 |
| Pop_A02G012299 | 2.225001206 | 0.013563496 | WRKY22 |
| Pop_A03G019779 | 2.390109471 | 0.001165373 | WRKY75 |
Table 2 Functional analysis of transcription factor-related DEGs
| 基因ID Gene ID | log2 (CL2/WT0) | Q value (WT0-vs-CL2) | 描述 Description |
|---|---|---|---|
| Pop_A01G003964 | 5.769956725 | 1.32E-41 | ERF1A |
| Pop_A01G032260 | 5.649644852 | 1.26E-05 | ERF21 |
| Pop_A01G059811 | 5.539774145 | 5.91E-07 | ERF61 |
| Pop_A01G060003 | 7.0861912 | 1.99E-09 | DREB1E |
| Pop_A04G018519 | 3.029867059 | 3.13E-06 | bHLH162 |
| Pop_A06G061940 | 2.433933971 | 0.032353534 | ORG2 |
| Pop_A14G044066 | 2.36624488 | 0.001496264 | bZip53 |
| Pop_A08G063663 | 2.588359391 | 4.82E-13 | CDF2 |
| Pop_G19G009429 | 2.356323627 | 6.09E-16 | CDF3 |
| Pop_G09G015087 | 2.033526993 | 0.003029387 | ZAT10 |
| Pop_A16G090138 | 3.342912788 | 3.50E-13 | PHL7 |
| Pop_G10G047973 | 6.429548834 | 0.001027136 | ORR25 |
| Pop_A09G014447 | 2.372666749 | 6.59E-06 | SCR3 |
| Pop_A06G062082 | 6.325653816 | 0.001158459 | HSF4b |
| Pop_A16G055308 | 2.667183627 | 5.18E-06 | HSF3 |
| Pop_A02G066179 | 5.110284639 | 8.26E-05 | MYB73 |
| Pop_A07G011878 | 2.013444428 | 1.37E-12 | MYB44 |
| Pop_A08G063813 | 2.04957107 | 0.013470953 | MYB108 |
| Pop_A14G000372 | 3.81186488 | 1.23E-08 | MYB44 |
| Pop_A15G064557 | 4.679506153 | 1.94E-16 | MYB62 |
| Pop_A04G019588 | 2.206753814 | 6.93E-06 | NAC73 |
| Pop_A04G026457 | 3.711825186 | 0.000111049 | TCP13 |
| Pop_A01G059075 | 3.408636654 | 6.29E-05 | WRKY28 |
| Pop_A01G080041 | 3.413796772 | 6.86E-30 | WRKY40 |
| Pop_A02G012299 | 2.225001206 | 0.013563496 | WRKY22 |
| Pop_A03G019779 | 2.390109471 | 0.001165373 | WRKY75 |
| 基因ID Gene ID | log2(CL12/WT0) | Q value | 描述 Description |
|---|---|---|---|
| Pop_A02G023792 | 1.224720934 | 1.09E-10 | GAUT8 |
| Pop_G02G015804 | 1.084322434 | 1.96E-08 | GAUT8 |
| Pop_G18G078711 | 1.659192116 | 3.08E-11 | CesA1 |
| Pop_A18G012937 | 1.547823248 | 7.46E-12 | CesA1 |
| Pop_A03G049677 | 2.0312488 | 1.97E-08 | PAE7 |
| Pop_A05G016925 | 1.624518546 | 3.92E-06 | PAE12 |
| Pop_G03G021316 | 1.771860445 | 3.93E-08 | PAE7 |
| Pop_G05G032831 | 1.404050162 | 4.41E-07 | PAE12 |
| Pop_G03G011242 | 1.373107821 | 4.15E-14 | PT |
| Pop_A02G023780 | 2.640491502 | 1.58E-21 | XTH2 |
| Pop_A19G055144 | 0.555458286 | 0.19872077 | XTH9 |
| Pop_G01G002830 | 2.869514357 | 0.070436159 | XTH27 |
| Pop_G02G015923 | 3.12618234 | 4.66E-63 | XTH21 |
| Pop_A01G074711 | 2.335379968 | 0.044535379 | XTHA |
| Pop_G19G084027 | 0.768927412 | 0.025571895 | XTH9 |
| Pop_G06G075982 | 1.258093365 | 0.194655673 | XTH6/TCH4 |
Table 3 Functional analysis of cell wall development-related DEGs
| 基因ID Gene ID | log2(CL12/WT0) | Q value | 描述 Description |
|---|---|---|---|
| Pop_A02G023792 | 1.224720934 | 1.09E-10 | GAUT8 |
| Pop_G02G015804 | 1.084322434 | 1.96E-08 | GAUT8 |
| Pop_G18G078711 | 1.659192116 | 3.08E-11 | CesA1 |
| Pop_A18G012937 | 1.547823248 | 7.46E-12 | CesA1 |
| Pop_A03G049677 | 2.0312488 | 1.97E-08 | PAE7 |
| Pop_A05G016925 | 1.624518546 | 3.92E-06 | PAE12 |
| Pop_G03G021316 | 1.771860445 | 3.93E-08 | PAE7 |
| Pop_G05G032831 | 1.404050162 | 4.41E-07 | PAE12 |
| Pop_G03G011242 | 1.373107821 | 4.15E-14 | PT |
| Pop_A02G023780 | 2.640491502 | 1.58E-21 | XTH2 |
| Pop_A19G055144 | 0.555458286 | 0.19872077 | XTH9 |
| Pop_G01G002830 | 2.869514357 | 0.070436159 | XTH27 |
| Pop_G02G015923 | 3.12618234 | 4.66E-63 | XTH21 |
| Pop_A01G074711 | 2.335379968 | 0.044535379 | XTHA |
| Pop_G19G084027 | 0.768927412 | 0.025571895 | XTH9 |
| Pop_G06G075982 | 1.258093365 | 0.194655673 | XTH6/TCH4 |
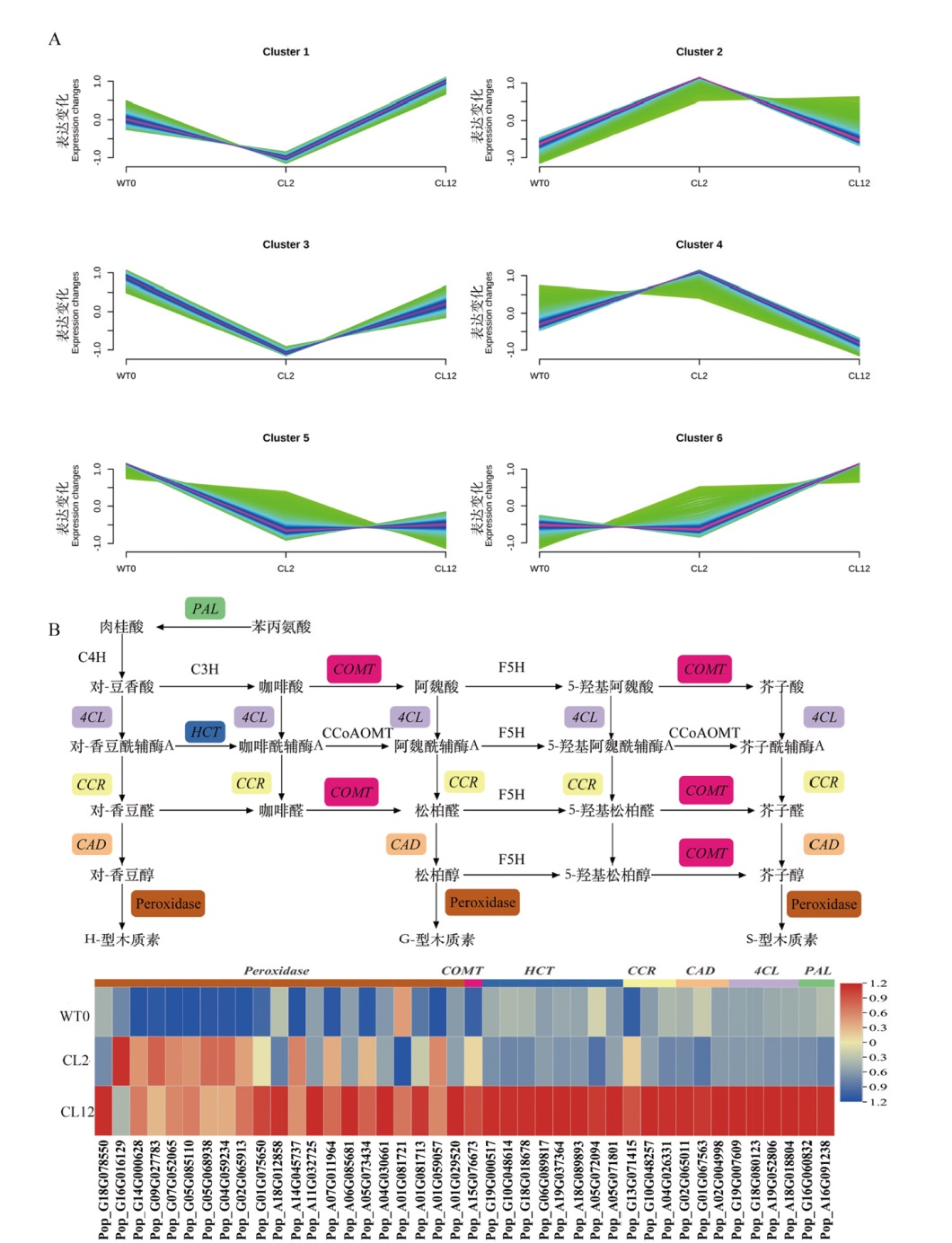
Fig. 4 DEGs temporal clustering and schematic representation of the phenylpropanoid pathway A: DEGs temporal clustering. B: Schematic representation of the phenylpropanoid pathway(Black boxes indicate genes encoding enzymes that are differentially expressed). WT0: Control. CL2:TSA treatment for 2 h. CL12:TSA treatment for 12 h
| 基因ID Gene ID | log2(CL12/WT0) | Q value | 描述 Description |
|---|---|---|---|
| Pop_A19G052806 | 2.403845576 | 4.60E-20 | 4CL2 |
| Pop_A18G018804 | 2.667476561 | 2.32E-10 | 4CL4 |
| Pop_G19G007609 | 3.667350303 | 2.16E-23 | 4CL2 |
| Pop_G18G080123 | 2.367966412 | 1.40E-07 | 4CL4 |
| Pop_A02G004998 | 3.561801582 | 7.11E-17 | CAD3 |
| Pop_G01G067563 | 1.220128387 | 0.02149788 | CAD9 |
| Pop_G02G065011 | 2.056960752 | 2.93E-12 | CAD3 |
| Pop_G10G048257 | 2.444394374 | 5.10E-14 | CCR3 |
| Pop_A04G026331 | 1.602900842 | 0.002526309 | CCR10 |
| Pop_G13G071415 | 3.465894044 | 6.89E-07 | CCR2 |
| Pop_A15G076673 | 3.006325753 | 1.29E-06 | COMT3 |
| Pop_A18G089893 | 2.322128861 | 0.000122309 | HCT |
| Pop_G06G089817 | 4.974228282 | 6.97E-05 | HCT |
| Pop_A19G037364 | 2.298057613 | 0.000168854 | HCT |
| Pop_G10G048614 | 2.130717747 | 1.18E-08 | HCT |
| Pop_G18G018678 | 2.236704931 | 0.078108456 | HCT |
| Pop_A05G071801 | 3.065678506 | 3.14E-07 | HCT1 |
| Pop_G19G000517 | 1.836359474 | 2.75E-05 | HCT |
| Pop_A05G072094 | 1.301262613 | 0.407703359 | HCT |
| Pop_A16G091238 | 1.580246846 | 6.22E-05 | PAL |
| Pop_G16G060832 | 2.402572772 | 4.33E-12 | PAL |
| Pop_A01G029520 | 3.361672474 | 0.005884321 | Peroxidase |
| Pop_A14G045737 | 3.712025804 | 5.26E-06 | Peroxidase |
| Pop_G01G075650 | 2.826658953 | 0.006729353 | Peroxidase |
| Pop_G04G059234 | 1.493633494 | 5.56E-06 | Peroxidase |
| Pop_A07G011964 | 3.400742219 | 5.98E-09 | Peroxidase |
| Pop_G02G065913 | 2.501334426 | 0.013145367 | Peroxidase |
| Pop_A06G085681 | 2.902707699 | 0.003229047 | Peroxidase |
| Pop_A04G030661 | 2.73324656 | 1.12E-05 | Peroxidase |
| Pop_G07G052065 | 3.735543904 | 1.65E-06 | Peroxidase |
| Pop_G14G000628 | 2.779869353 | 0.006974024 | Peroxidase |
| Pop_G16G016129 | 0.785917676 | 0.479463762 | Peroxidase |
| Pop_G05G068938 | 1.80032676 | 1.21E-06 | Peroxidase |
| Pop_A01G059057 | 1.923239289 | 5.70E-28 | Peroxidase |
| Pop_G09G027783 | 1.931046589 | 6.19E-07 | Peroxidase |
| Pop_G05G085110 | 2.311933723 | 1.03E-11 | Peroxidase |
| Pop_A05G073434 | 2.266091449 | 1.44E-08 | Peroxidase |
| Pop_G18G078550 | 2.34410638 | 1.22E-13 | Peroxidase |
| Pop_A18G012858 | 1.673704695 | 3.66E-08 | Peroxidase |
| Pop_A01G081713 | 6.43779021 | 0.000215695 | Peroxidase |
| Pop_A01G081721 | 0.235613668 | 0.858752785 | Peroxidase |
| Pop_A11G032725 | 3.074623654 | 4.13E-15 | Peroxidase |
Table 4 Function analysis of DEGs involved in phenylpropanoid biosynthesis
| 基因ID Gene ID | log2(CL12/WT0) | Q value | 描述 Description |
|---|---|---|---|
| Pop_A19G052806 | 2.403845576 | 4.60E-20 | 4CL2 |
| Pop_A18G018804 | 2.667476561 | 2.32E-10 | 4CL4 |
| Pop_G19G007609 | 3.667350303 | 2.16E-23 | 4CL2 |
| Pop_G18G080123 | 2.367966412 | 1.40E-07 | 4CL4 |
| Pop_A02G004998 | 3.561801582 | 7.11E-17 | CAD3 |
| Pop_G01G067563 | 1.220128387 | 0.02149788 | CAD9 |
| Pop_G02G065011 | 2.056960752 | 2.93E-12 | CAD3 |
| Pop_G10G048257 | 2.444394374 | 5.10E-14 | CCR3 |
| Pop_A04G026331 | 1.602900842 | 0.002526309 | CCR10 |
| Pop_G13G071415 | 3.465894044 | 6.89E-07 | CCR2 |
| Pop_A15G076673 | 3.006325753 | 1.29E-06 | COMT3 |
| Pop_A18G089893 | 2.322128861 | 0.000122309 | HCT |
| Pop_G06G089817 | 4.974228282 | 6.97E-05 | HCT |
| Pop_A19G037364 | 2.298057613 | 0.000168854 | HCT |
| Pop_G10G048614 | 2.130717747 | 1.18E-08 | HCT |
| Pop_G18G018678 | 2.236704931 | 0.078108456 | HCT |
| Pop_A05G071801 | 3.065678506 | 3.14E-07 | HCT1 |
| Pop_G19G000517 | 1.836359474 | 2.75E-05 | HCT |
| Pop_A05G072094 | 1.301262613 | 0.407703359 | HCT |
| Pop_A16G091238 | 1.580246846 | 6.22E-05 | PAL |
| Pop_G16G060832 | 2.402572772 | 4.33E-12 | PAL |
| Pop_A01G029520 | 3.361672474 | 0.005884321 | Peroxidase |
| Pop_A14G045737 | 3.712025804 | 5.26E-06 | Peroxidase |
| Pop_G01G075650 | 2.826658953 | 0.006729353 | Peroxidase |
| Pop_G04G059234 | 1.493633494 | 5.56E-06 | Peroxidase |
| Pop_A07G011964 | 3.400742219 | 5.98E-09 | Peroxidase |
| Pop_G02G065913 | 2.501334426 | 0.013145367 | Peroxidase |
| Pop_A06G085681 | 2.902707699 | 0.003229047 | Peroxidase |
| Pop_A04G030661 | 2.73324656 | 1.12E-05 | Peroxidase |
| Pop_G07G052065 | 3.735543904 | 1.65E-06 | Peroxidase |
| Pop_G14G000628 | 2.779869353 | 0.006974024 | Peroxidase |
| Pop_G16G016129 | 0.785917676 | 0.479463762 | Peroxidase |
| Pop_G05G068938 | 1.80032676 | 1.21E-06 | Peroxidase |
| Pop_A01G059057 | 1.923239289 | 5.70E-28 | Peroxidase |
| Pop_G09G027783 | 1.931046589 | 6.19E-07 | Peroxidase |
| Pop_G05G085110 | 2.311933723 | 1.03E-11 | Peroxidase |
| Pop_A05G073434 | 2.266091449 | 1.44E-08 | Peroxidase |
| Pop_G18G078550 | 2.34410638 | 1.22E-13 | Peroxidase |
| Pop_A18G012858 | 1.673704695 | 3.66E-08 | Peroxidase |
| Pop_A01G081713 | 6.43779021 | 0.000215695 | Peroxidase |
| Pop_A01G081721 | 0.235613668 | 0.858752785 | Peroxidase |
| Pop_A11G032725 | 3.074623654 | 4.13E-15 | Peroxidase |

Fig. 6 Phenotypic analysis of poplar plants treated with TSA A: Untreated(left)and TSA-treated(right)poplar plants. B: Statistics of plant heights. C-N: Paraffin section analysis of poplar stem cross-sections. C-E: The 18th stem segment of control plants. F-H: The 18th stem segment of TSA-treated plants. I-K: The 16th stem segment of control plants. L-N: The 16th stem segment of TSA-treated plants; C, F, I, L scale bar = 200 μm; D, G, J, M scale bar=100 μm; E, H, K, N scale bar=50 μm. O: Statistics of plant stem diameter. P: Statistics of plant xylem thickness. *P<0.05, **P<0.01

Fig. 7 SEM analysis of the xylem phenotype in poplar A-D: SEM analysis of the xylem structure in poplar. A, B: Untreated group; C, D: TSA-treated group. A, C scale bars = 500 μm; B, D scale bars =100 μm. E: Statistics of vessel area. F: Statistics of vessel number
| [1] | 张冰, 夏德安, 马旭俊. 组蛋白去乙酰化酶在杨树根再生和生长中的功能[J]. 江苏农业科学, 2017, 45(5): 40-43. |
| Zhang B, Xia DA, Ma XJ. Function of histone deacetylase in regeneration and growth of Yang Shugen[J]. Jiangsu Agric Sci, 2017, 45(5): 40-43. | |
| [2] |
Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention[J]. Oncogene, 2007, 26(37): 5310-5318.
doi: 10.1038/sj.onc.1210599 pmid: 17694074 |
| [3] |
Dangl M, Brosch G, Haas H, et al. Comparative analysis of HD2 type histone deacetylases in higher plants[J]. Planta, 2001, 213(2): 280-285.
pmid: 11469594 |
| [4] | Servet C, Silva NCE, et al. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis[J]. Mol Plant, 2010, 3(4): 670-677. |
| [5] | 李淑娟, 张超, 张彦妮, 等. 毛果杨HDAC基因家族序列及其表达分析[J]. 西北农林科技大学学报: 自然科学版, 2015, 43(3): 63-76. |
| Li SJ, Zhang C, Zhang YN, et al. Sequence and gene expression of histone deacetylases(HDAC)gene family of Populus trichocar-pa[J]. J Northwest A F Univ Nat Sci Ed, 2015, 43(3): 63-76. | |
| [6] |
Chen XS, Ding AB, et al. Functions and mechanisms of plant histone deacetylases[J]. Sci China Life Sci, 2020, 63(2): 206-216.
doi: 10.1007/s11427-019-1587-x pmid: 31879846 |
| [7] |
Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19[J]. Development, 2012, 139(22): 4180-4190.
doi: 10.1242/dev.085407 pmid: 23034631 |
| [8] | Yu CW, Liu XC, Luo M, et al. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidop-sis[J]. Plant Physiol, 2011, 156(1): 173-184. |
| [9] | Yu CW, Chang KY, Wu KQ. Genome-wide analysis of gene regulatory networks of the FVE-HDA6-FLD complex in Arabidopsis[J]. Front Plant Sci, 2016, 7: 555. |
| [10] | Kang MJ, Jin HS, Noh YS, et al. Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis[J]. New Phytol, 2015, 206(1): 281-294. |
| [11] | Luo M, Yu CW, Chen FF, et al. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis[J]. PLoS Genet, 2012, 8(12): e1003114. |
| [12] | Chen XS, Lu L, Mayer KS, et al. POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidop-sis[J]. eLife, 2016, 5: e17214. |
| [13] | Li HC, Torres-Garcia J, Latrasse D, et al. Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE2 expression to control Arabidopsis root meristem cell number[J]. Plant Cell, 2017, 29(9): 2183-2196. |
| [14] | Chung PJ, Kim YS, et al. The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice[J]. Plant J, 2009, 59(5): 764-776. |
| [15] |
Luo M, Cheng K, et al. Plant responses to abiotic stress regulated by histone deacetylases[J]. Front Plant Sci, 2017, 8: 2147.
doi: 10.3389/fpls.2017.02147 pmid: 29326743 |
| [16] | Lee HG, Seo PJ. MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis[J]. Nat Commun, 2019, 10(1): 1713. |
| [17] | Shen Y, Lei TT, Cui XY, et al. Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature[J]. Plant J, 2019, 100(5): 991-1006. |
| [18] | To TK, Nakaminami K, Kim JM, et al. Arabidopsis HDA6 is required for freezing tolerance[J]. Biochem Biophys Res Commun, 2011, 406(3): 414-419. |
| [19] | Dai XF, Zhai R, Lin JJ, et al. Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa[J]. Nat Plants, 2023, 9(1): 96-111. |
| [20] | Zhang YZ, Yin B, Zhang JX, et al. Histone deacetylase HDT1 is involved in stem vascular development in Arabidopsis[J]. Int J Mol Sci, 2019, 20(14): 3452. |
| [21] | Hirai R, Wang SM, Demura T, et al. Histone deacetylation controls xylem vessel cell differentiation via transcriptional regulation of a transcription repressor complex OFP1/4-MYB75-KNAT7-BLH6[J]. Front Plant Sci, 2022, 12: 825810. |
| [22] | Li S, Lin YCJ, et al. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa[J]. Plant Cell, 2019, 31(3): 663-686. |
| [23] | Nowak K, Morończyk J, Grzyb M, et al. miR172 regulates WUS during somatic embryogenesis in Arabidopsis via AP2[J]. Cells, 2022, 11(4): 718. |
| [24] |
Zheng Y, Ge JY, Bao C, et al. Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response[J]. Mol Plant, 2020, 13(4): 598-611.
doi: S1674-2052(19)30408-3 pmid: 31891777 |
| [25] | Gao SY, Yin MX, Xu MY, et al. Transcription factors PuPRE6/PuMYB12 and histone deacetylase PuHDAC9-like regulate sucrose levels in pear[J]. Plant Physiol, 2024, 194(3): 1577-1592. |
| [26] | 李慧, 郭晓蕊, 刘雅琳, 等. 木材形成过程中次生壁沉积和细胞程序性死亡的分子调控机制[J]. 中国科学: 生命科学, 2020, 50(2): 123-135. |
| Li H, Guo XR, Liu YL, et al. The molecular mechanism in secondary wall deposition and programmed cell death of wood formation[J]. Sci Sin Vitae, 2020, 50(2): 123-135. | |
| [27] | Shahin L, Zhang L, Mohnen D, et al. Insights into pectin O-acetylation in the plant cell wall: structure, synthesis, and modification[J]. Cell Surf, 2023, 9: 100099. |
| [28] |
Fan NN, Su LT, Lv AM, et al. PECTIN ACETYLESTERASE12 regulates shoot branching via acetic acid and auxin accumulation in alfalfa shoots[J]. Plant Physiol, 2024, 195(1): 518-533.
doi: 10.1093/plphys/kiae071 pmid: 38365203 |
| [29] | Hrmova M, Stratilová B, et al. Broad specific xyloglucan: xyloglucosyl transferases are formidable players in the re-modelling of plant cell wall structures[J]. Int J Mol Sci, 2022, 23(3): 1656. |
| [30] |
Zhong RQ, Cui DT, Ye ZH. Secondary cell wall biosynthesis[J]. New Phytol, 2019, 221(4): 1703-1723.
doi: 10.1111/nph.15537 pmid: 30312479 |
| [31] | 李金花, 张绮纹, 牛正田, 等. 木质素生物合成及其基因调控的研究进展[J]. 世界林业研究, 2007, 20(1): 29-37. |
| Li JH, Zhang QW, Niu ZT, et al. Advances in study of lignin biosynthesis and genetic engineering modification[J]. World For Res, 2007, 20(1): 29-37. |
| [1] | PEI Xu-juan, DI Jing-yi, LIU Hao, GAO Wei-xia. Exploration of Regulatory Elements for Hyaluronic Acid Molecular Weight in Streptococcus zooepidemicus via Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(1): 347-356. |
| [2] | NIE Zhu-xin, GUO Jin, QIAO Zi-yang, LI Wei-wei, ZHANG Xue-yan, LIU Chun-yang, WANG Jing. Transcriptome Analysis of the Anthocyanin Biosynthesis in the Fruit Development Processes of Lycium ruthenicum Murr. [J]. Biotechnology Bulletin, 2024, 40(8): 106-117. |
| [3] | ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata [J]. Biotechnology Bulletin, 2024, 40(8): 142-151. |
| [4] | LIAO Yang-mei, ZHAO Guo-chun, WENG Xue-huang, JIA Li-ming, CHEN Zhong. Transcriptome Sequencing of Male Sterile Buds at Different Developmental Stages in Sapindus mukorossi ‘Qirui’ [J]. Biotechnology Bulletin, 2024, 40(7): 197-206. |
| [5] | GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean [J]. Biotechnology Bulletin, 2024, 40(7): 137-149. |
| [6] | BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility [J]. Biotechnology Bulletin, 2024, 40(6): 134-142. |
| [7] | WU Di, YOU Xiao-feng, ZHENG Yi-zheng, LIN Nan, ZHANG Yan-yan, WEI Yi-cong. Analysis of Endogenous Hormone Regulation Mechanism for Carotenoid Synthesis in Sarcandra glabra [J]. Biotechnology Bulletin, 2024, 40(5): 203-214. |
| [8] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [9] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [10] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [11] | XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality [J]. Biotechnology Bulletin, 2024, 40(3): 215-228. |
| [12] | LIANG Jia-lin, ZHAO Shuang, LI Xing-er, ZHAO Cheng-zhou, LI Ping. Identification of Aux/IAA Gene Family in Corydalis hendersonii Hemsl. and Analysis on Their Expression Pattern under UVB Treatment [J]. Biotechnology Bulletin, 2024, 40(12): 182-192. |
| [13] | LIU Shu-tong, WU Sheng, TAN Yi-yang, WANG De-pei, XUE Xian-li. Differential Analysis of Key Genes Involved in the Accumulation of L-malic Acid by Aspergillus niger Fermentation [J]. Biotechnology Bulletin, 2024, 40(12): 227-238. |
| [14] | LIANG Wan-feng, ZENG Jing-jing, HU Ruo-qun, CAO Jia-yu, ZHENG Tao, LI Luan, QIU Ming-yue, LIANG Xiao-ying, CHEN Ying. Transcriptional and Metabolomic Analysis of Carotenoid Accumulation in Anoectochilus roxburghii during Different Growth Periods [J]. Biotechnology Bulletin, 2024, 40(10): 262-274. |
| [15] | LI Ming-kun, BI Mei-ying, ZHANG Tian-hang, WU Xiang-yu, YANG Pei-ru, YING Ming. Restoration of Agricultural Function of Rhizobacteria by UgRNA/Cas9 Multi-gene Editing [J]. Biotechnology Bulletin, 2024, 40(10): 275-287. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||