








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 12-26.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0056
Previous Articles Next Articles
LYU Yue1( ), ZHANG Jie-wei2(
), ZHANG Jie-wei2( ), WANG Bo1(
), WANG Bo1( )
)
Received:2025-01-13
Online:2025-06-26
Published:2025-06-30
Contact:
ZHANG Jie-wei, WANG Bo
E-mail:lvyue19819083151@163.com;jwzhang919@163.com;wangbo@ytu.edu.cn
LYU Yue, ZHANG Jie-wei, WANG Bo. Research Progress in RNA Binding Proteins in Plant Disease Resistance[J]. Biotechnology Bulletin, 2025, 41(6): 12-26.

Fig. 1 Topology diagram of common RNA-binding domainsA: Schematic diagram of RNA recognition motif topology; B: schematic diagram of K-homology domain topology; C: schematic diagram of zinc-finger domain topology; D: schematic diagram of double-stranded RNA binding domain topology

Fig. 2 Diagram of AS eventThe red rectangle, purple rectangle, yellow rectangle, and blue rectangle indicate exon 1 to 4, respectively; the light blue rectangle indicates exon fragments that undergo 5' alternative splicing or 3' alternative splicing; the thick gray line indicates the intron; the solid black line indicates the first AS (alternative splicing) mode; the dashed black line indicates the second AS mode; and the arrow points to the AS product
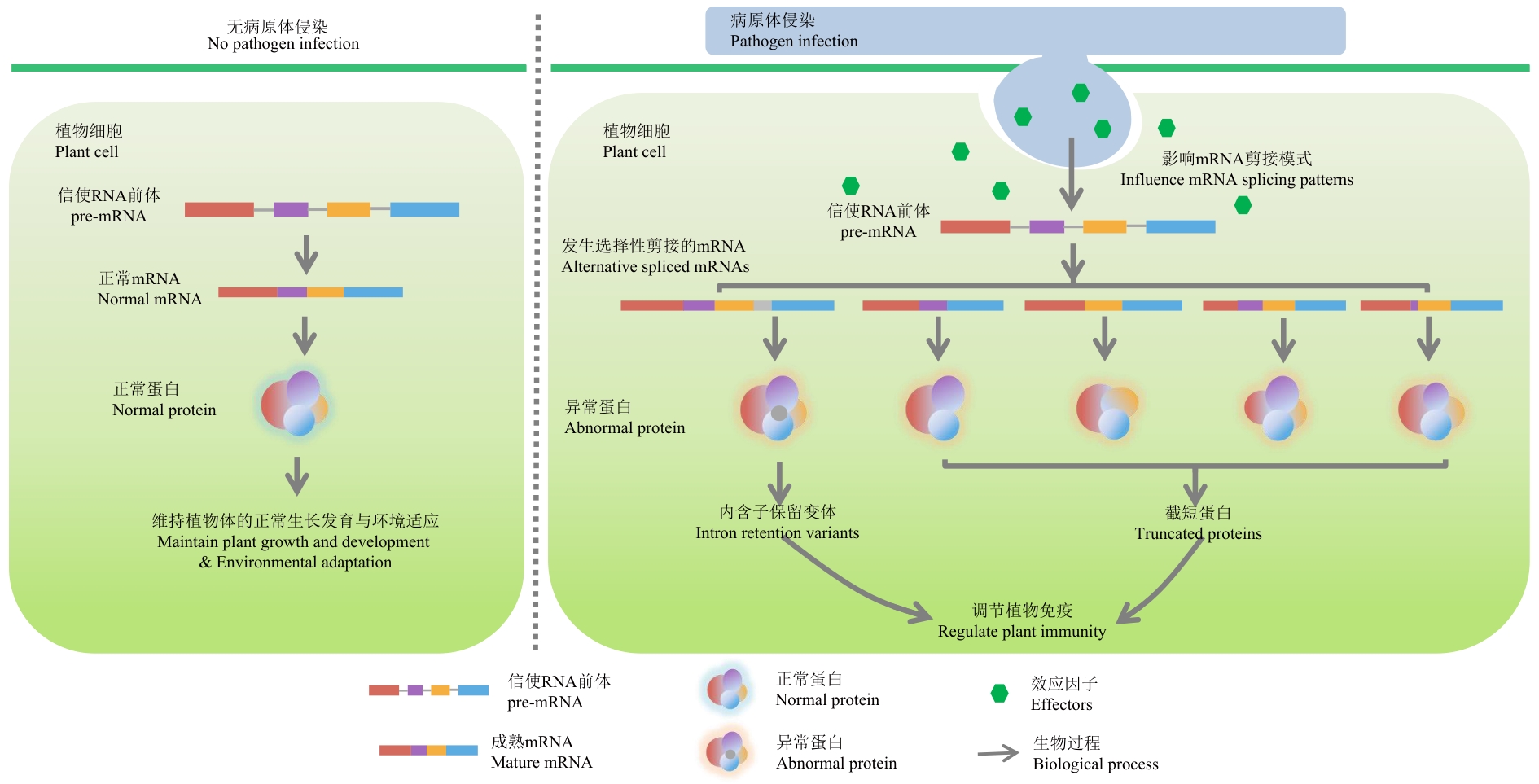
Fig. 3 Alternative splicing patterns of RBPs-participating genesThe green hexagon symbolizes the effect factor. Under natural conditions, plant pre-mRNAs undergo constitutive splicing to generate full-length mature mRNAs, which are translated into functional proteins essential for normal growth, development, and adaptation to environment. Upon pathogenic infection, pathogens release effector proteins into plant cells that either act as RNA-binding proteins (RBPs) or target host RBPs to perturb AS. This interference leads to the production of intron-retained variants or truncated proteins, thereby disrupting plant immune regulation

Fig. 4 Schematic diagram of RBPs-regulating mechanismA: Schematic diagram of RBPs participating in plant disease resistance mechanism through AS of genes. B: Schematic diagram of RBPs participating in plant disease resistance through protein translation. C: Schematic diagram of RBPs participating in plant disease resistance through APA. D: Schematic diagram of RBPs participating in plant disease resistance by regulating mRNA stability. E: Schematic diagram of RNA modification involved in plant disease resistance

Fig. 5 Schematic diagram of mRNA stability regulation mechanismGreen hexagons denote effect factors, blue circles represent RBPs, blue ovals represent degraded RBPs, wavy lines symbolize various mRNAs encoding proteins associated with plant immunity, arrows signify biological processes. Under natural conditions, plant RBPs bind to mRNA, stabilizing it and ensuring the smooth execution of translation and other biological processes, thereby maintaining normal growth, development, and environmental adaptation in plants. Upon pathogen infection, pathogens release effectors into plant cells. These effectors may either down-regulate the expression of RBPs through fungal stimulation or degradation by interaction with effectors. The reduced abundance of RBPs leads to decreased mRNA stability, which in turn affects the expression of immune-related proteins and ultimately compromises the plant’s resistance to pathogens
| 1 | 王郑雷. 转录组和代谢组联合分析椭圆叶花锚响应海拔的关键代谢通路 [D]. 西宁: 青海师范大学, 2023. |
| Wang ZL. Transcriptome and metabonomics joint analysis of key metabolic pathways of Anopheles elliptica in response to altitude [D]. Xining: Qinghai Normal University, 2023. | |
| 2 | Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation [J]. FEBS Lett, 2008, 582(14): 1977-1986. |
| 3 | Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs [J]. Mol Cell, 2014, 54(4): 547-558. |
| 4 | Singh G, Pratt G, Yeo GW, et al. The clothes make the mRNA: past and present trends in mRNP fashion [J]. Annu Rev Biochem, 2015, 84: 325-354. |
| 5 | Corley M, Burns MC, Yeo GW. How RNA-binding proteins interact with RNA: molecules and mechanisms [J]. Mol Cell, 2020, 78(1): 9-29. |
| 6 | Gehring NH, Wahle E, Fischer U. Deciphering the mRNP code: rna-bound determinants of post-transcriptional gene regulation [J]. Trends Biochem Sci, 2017, 42(5): 369-382. |
| 7 | Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function [J]. Nat Rev Mol Cell Biol, 2007, 8(6): 479-490. |
| 8 | Zhang Y, Xu Y, Skaggs TH, et al. Plant phase extraction: a method for enhanced discovery of the RNA-binding proteome and its dynamics in plants [J]. Plant Cell, 2023, 35(8): 2750-2772. |
| 9 | Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins [J]. Nat Rev Genet, 2014, 15(12): 829-845. |
| 10 | Chen Y, Varani G. Engineering RNA-binding proteins for biology [J]. FEBS J, 2013, 280(16): 3734-3754. |
| 11 | Fan S, Zhang Y, Zhu SB, et al. Plant RNA-binding proteins: phase separation dynamics and functional mechanisms underlying plant development and stress responses [J]. Mol Plant, 2024, 17(4): 531-551. |
| 12 | 闫宗运. 拟南芥中含有CCCH锌指域和KH域的蛋白调控成花与衰老的机理 [D]. 北京: 中国农业大学, 2018. |
| Yan ZY. Mechanism of regulating flowering and senescence by proteins containing CCCH zinc finger domain and KH domain in Arabidopsis thaliana [D]. Beijing: China Agricultural University, 2018. | |
| 13 | 侯毅枫. RNA结合蛋白HLP1与HLP2调控拟南芥开花时间的分子机理研究 [D]. 泰安: 山东农业大学, 2016. |
| Hou YF. Molecular mechanism of RNA binding proteins HLP1 and HLP2 regulating flowering time of Arabidopsis thaliana [D]. Tai’an: Shandong Agricultural University, 2016. | |
| 14 | Zhang Y, Fan S, Hua CM, et al. Phase separation of HRLP regulates flowering time in Arabidopsis [J]. Sci Adv, 2022, 8(25): eabn5488. |
| 15 | Ma LQ, Yang YF, Wang YQ, et al. SlRBP1 promotes translational efficiency via SleIF4A2 to maintain chloroplast function in tomato [J]. Plant Cell, 2022, 34(7): 2747-2764. |
| 16 | Lorković ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana [J]. Nucleic Acids Res, 2002, 30(3): 623-635. |
| 17 | Duque P. A role for SR proteins in plant stress responses [J]. Plant Signal Behav, 2011, 6(1): 49-54. |
| 18 | Lorković ZJ. Role of plant RNA-binding proteins in development, stress response and genome organization [J]. Trends Plant Sci, 2009, 14(4): 229-236. |
| 19 | Duthie G, Crozier A. Plant-derived phenolic antioxidants [J]. Curr Opin Clin Nutr Metab Care, 2000, 3(6): 447-451. |
| 20 | 陈玲. 新型RNA结合蛋白SKRP负调控植物对疫霉菌的免疫机制研究 [D]. 南京: 南京农业大学, 2021. |
| Chen L. A novel RNA binding protein SKRP negatively regulates the immune mechanism of plants to Phytophthora infestans [D]. Nanjing: Nanjing Agricultural University, 2021. | |
| 21 | Lyons R, Iwase A, Gänsewig T, et al. The RNA-binding protein FPA regulates flg22-triggered defense responses and transcription factor activity by alternative polyadenylation [J]. Sci Rep, 2013, 3: 2866. |
| 22 | Zhang JC, Zheng HY, Li YW, et al. Coexpression network analysis of the genes regulated by two types of resistance responses to powdery mildew in wheat [J]. Sci Rep, 2016, 6: 23805. |
| 23 | Wang RL, Wang Y, He DD, et al. Responses of plant immune system and rhizosphere soil microbiome to the elicitor BAR11 in Arabidopsis thaliana [J]. Sci Total Environ, 2024, 914: 169920. |
| 24 | Ma FL, Yang XF, Shi ZY, et al. Novel crosstalk between ethylene- and jasmonic acid-pathway responses to a piercing-sucking insect in rice [J]. New Phytol, 2020, 225(1): 474-487. |
| 25 | Xu J, Wang XJ, Zu HY, et al. Molecular dissection of rice phytohormone signaling involved in resistance to a piercing-sucking herbivore [J]. New Phytol, 2021, 230(4): 1639-1652. |
| 26 | Yuan MH, Jiang ZY, Bi GZ, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity [J]. Nature, 2021, 592(7852): 105-109. |
| 27 | Ngou BPM, Ding PT, Jones JDG. Thirty years of resistance: Zig-zag through the plant immune system [J]. Plant Cell, 2022, 34(5): 1447-1478. |
| 28 | Staiger D, Brown JWS. Alternative splicing at the intersection of biological timing, development, and stress responses [J]. Plant Cell, 2013, 25(10): 3640-3656. |
| 29 | Syed NH, Kalyna M, Marquez Y, et al. Alternative splicing in plants—coming of age [J]. Trends Plant Sci, 2012, 17(10): 616-623. |
| 30 | Chaudhary S, Khokhar W, Jabre I, et al. Alternative splicing and protein diversity: plants versus animals [J]. Front Plant Sci, 2019, 10: 708. |
| 31 | Marquez Y, Brown JWS, Simpson C, et al. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis [J]. Genome Res, 2012, 22(6): 1184-1195. |
| 32 | Abulfaraj AA, Alhoraibi HM, Mariappan K, et al. Analysis of the Arabidopsis coilin mutant reveals a positive role of AtCOILIN in plant immunity [J]. Plant Physiol, 2022, 190(1): 745-761. |
| 33 | Zhang RX, Calixto CPG, Marquez Y, et al. A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing [J]. Nucleic Acids Res, 2017, 45(9): 5061-5073. |
| 34 | Ling ZH, Zhou WW, Baldwin IT, et al. Insect herbivory elicits genome-wide alternative splicing responses in Nicotiana attenuata [J]. Plant J, 2015, 84(1): 228-243. |
| 35 | Bazin J, Mariappan K, Jiang YH, et al. Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis [J]. PLoS Pathog, 2020, 16(4): e1008401. |
| 36 | Howard BE, Hu QW, Babaoglu AC, et al. High-throughput RNA sequencing of Pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants [J]. PLoS One, 2013, 8(10): e74183. |
| 37 | Huang J, Lu XY, Wu HW, et al. Phytophthora effectors modulate genome-wide alternative splicing of host mRNAs to reprogram plant immunity [J]. Mol Plant, 2020, 13(10): 1470-1484. |
| 38 | Jin LR, Li GL, Yu DZ, et al. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus Verticillium dahliae [J]. BMC Genomics, 2017, 18(1): 130. |
| 39 | Xu F, Xu SH, Wiermer M, et al. The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes [J]. Plant J, 2012, 70(6): 916-928. |
| 40 | Zhang ZB, Liu YN, Ding PT, et al. Splicing of receptor-like kinase-encoding SNC4 and CERK1 is regulated by two conserved splicing factors that are required for plant immunity [J]. Mol Plant, 2014, 7(12): 1766-1775. |
| 41 | Wu Z, Zhu DL, Lin XY, et al. RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis [J]. Plant Cell, 2016, 28(1): 55-73. |
| 42 | Fang Y, Chen J. Changing the script: a Phytophthora sojae effector hijacks RNA-binding proteins to reprogram plant immunity [J]. Plant Physiol, 2023, 191(2): 820-822. |
| 43 | Gui XM, Zhang P, Wang D, et al. Phytophthora effector PSR1 hijacks the host pre-mRNA splicing machinery to modulate small RNA biogenesis and plant immunity [J]. Plant Cell, 2022, 34(9): 3443-3459. |
| 44 | Gassmann W. Alternative splicing in plant defense [J]. Curr Top Microbiol Immunol, 2008, 326: 219-233. |
| 45 | Sánchez-Martín J, Widrig V, Herren G, et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins [J]. Nat Plants, 2021, 7(3): 327-341. |
| 46 | Sánchez-Martín J, Widrig V, Herren G, et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins [J]. Nat Plants, 2023, 9: 502-503. |
| 47 | Peng S, Guo DB, Guo Y, et al. Constitutive expresser of pathogenesis-related genes 5 is an RNA-binding protein controlling plant immunity via an RNA processing complex [J]. Plant Cell, 2022, 34(5): 1724-1744. |
| 48 | Zhang XN, Shi YF, Powers JJ, et al. Transcriptome analyses reveal SR45 to be a neutral splicing regulator and a suppressor of innate immunity in Arabidopsis thaliana [J]. BMC Genomics, 2017, 18(1): 772. |
| 49 | Xu SH, Zhang ZB, Jing BB, et al. Transportin-SR is required for proper splicing of resistance genes and plant immunity [J]. PLoS Genet, 2011, 7(6): e1002159. |
| 50 | Califice S, Baurain D, Hanikenne M, et al. A single ancient origin for prototypical serine/arginine-rich splicing factors [J]. Plant Physiol, 2012, 158(2): 546-560. |
| 51 | Qiao YL, Xia R, Zhai JX, et al. Small RNAs in plant immunity and virulence of filamentous pathogens [J]. Annu Rev Phytopathol, 2021, 59: 265-288. |
| 52 | Song XW, Li Y, Cao XF, et al. microRNAs and their regulatory roles in plant-environment interactions [J]. Annu Rev Plant Biol, 2019, 70: 489-525. |
| 53 | Xiong RY, Wu JX, Zhou YJ, et al. Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus [J]. Virology, 2009, 387(1): 29-40. |
| 54 | Wu JG, Yang ZR, Wang Y, et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA [J]. eLife, 2015, 4: e05733. |
| 55 | Du P, Wu JG, Zhang JY, et al. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors [J]. PLoS Pathog, 2011, 7(8): e1002176. |
| 56 | Lian S, Cho WK, Kim SM, et al. Time-course small RNA profiling reveals rice miRNAs and their target genes in response to rice stripe virus infection [J]. PLoS One, 2016, 11(9): e0162319. |
| 57 | Zheng LJ, Zhang C, Shi CN, et al. Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice [J]. PLoS Pathog, 2017, 13(10): e1006662. |
| 58 | Wang MH, Ma TL, Wang HX, et al. The RNA binding protein FgRbp1 regulates specific pre-mRNA splicing via interacting with U2AF23 in Fusarium [J]. Nat Commun, 2021, 12(1): 2661. |
| 59 | Huang J, Gu LF, Zhang Y, et al. An oomycete plant pathogen reprograms host pre-mRNA splicing to subvert immunity [J]. Nat Commun, 2017, 8(1): 2051. |
| 60 | Chen L, Xu ZH, Huang J, et al. Plant immunity suppressor SKRP encodes a novel RNA-binding protein that targets exon 3' end of unspliced RNA [J]. New Phytol, 2023, 240(4): 1467-1483. |
| 61 | Lee MW, Huffaker A, Crippen D, et al. Plant elicitor peptides promote plant defences against nematodes in soybean [J]. Mol Plant Pathol, 2018, 19(4): 858-869. |
| 62 | Huffaker A. Plant elicitor peptides in induced defense against insects [J]. Curr Opin Insect Sci, 2015, 9: 44-50. |
| 63 | Ruiz C, Nadal A, Montesinos E, et al. Novel Rosaceae plant elicitor peptides as sustainable tools to control Xanthomonas arboricola pv. pruni in Prunus spp [J]. Mol Plant Pathol, 2018, 19(2): 418-431. |
| 64 | Krol E, Mentzel T, Chinchilla D, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2 [J]. J Biol Chem, 2010, 285(18): 13471-13479. |
| 65 | Kadota Y, Sklenar J, Derbyshire P, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity [J]. Mol Cell, 2014, 54(1): 43-55. |
| 66 | Dressano K, Weckwerth PR, Poretsky E, et al. Dynamic regulation of Pep-induced immunity through post-translational control of defence transcript splicing [J]. Nat Plants, 2020, 6(8): 1008-1019. |
| 67 | Ma LQ, Cheng K, Li JY, et al. Roles of plant Glycine-rich RNA-binding proteins in development and stress responses [J]. Int J Mol Sci, 2021, 22(11): 5849. |
| 68 | Zicola J. From the archives: regulation of mRNA translation, epigenetic machinery components, and processing of a composite large subunit ribosomal RNA [J]. Plant Cell, 2023, 35(7): 2429-2430. |
| 69 | Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms [J]. RNA, 2002, 8(3): 265-278. |
| 70 | Fujisaki K, Ishikawa M. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication [J]. Virology, 2008, 380(2): 402-411. |
| 71 | Sugiura R, Kita A, Shimizu Y, et al. Feedback regulation of MAPK signalling by an RNA-binding protein [J]. Nature, 2003, 424(6951): 961-965. |
| 72 | Jeong RD, Chandra-Shekara AC, Kachroo A, et al. HRT-mediated hypersensitive response and resistance to Turnip crinkle virus in Arabidopsis does not require the function of TIP, the presumed guardee protein [J]. Mol Plant Microbe Interact, 2008, 21(10): 1316-1324. |
| 73 | Marrocco K, Criqui MC, Zervudacki J, et al. APC/C-mediated degradation of dsRNA-binding protein 4 (DRB4) involved in RNA silencing [J]. PLoS One, 2012, 7(4): e35173. |
| 74 | Lim GH, Hoey T, Zhu SF, et al. COP1, a negative regulator of photomorphogenesis, positively regulates plant disease resistance via double-stranded RNA binding proteins [J]. PLoS Pathog, 2018, 14(3): e1006894. |
| 75 | Singh A, Pandey A, Srivastava AK, et al. Plant protein phosphatases 2C: from genomic diversity to functional multiplicity and importance in stress management [J]. Crit Rev Biotechnol, 2016, 36(6): 1023-1035. |
| 76 | Ai G, Li T, Zhu H, et al. BPL3 binds the long non-coding RNA nalncFL7 to suppress FORKED-LIKE7 and modulate HAI1-mediated MPK3/6 dephosphorylation in plant immunity [J]. Plant Cell, 2023, 35(1): 598-616. |
| 77 | Wang LJ, Pei ZY, Tian YC, et al. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation [J]. Mol Plant Microbe Interact, 2005, 18(5): 375-384. |
| 78 | Liu JL, Park CH, He F, et al. The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice [J]. PLoS Pathog, 2015, 11(2): e1004629. |
| 79 | Zhang F, Fang H, Wang M, et al. APIP5 functions as a transcription factor and an RNA-binding protein to modulate cell death and immunity in rice [J]. Nucleic Acids Res, 2022, 50(9): 5064-5079. |
| 80 | Li Y, Zhang RR, Wu Y, et al. TaRBP1 stabilizes TaGLTP and negatively regulates stripe rust resistance in wheat [J]. Mol Plant Pathol, 2023, 24(10): 1205-1219. |
| 81 | Wang WD, Nie JJ, Lv LQ, et al. A Valsa mali effector protein 1 targets apple (Malus domestica) pathogenesis-related 10 protein to promote virulence [J]. Front Plant Sci, 2021, 12: 741342. |
| 82 | Wang WD, Wang SL, Gong W, et al. Valsa mali secretes an effector protein VmEP1 to target a K homology domain-containing protein for virulence in apple [J]. Mol Plant Pathol, 2022, 23(11): 1577-1591. |
| 83 | Turrà D, Segorbe D, Di Pietro A. Protein kinases in plant-pathogenic fungi: conserved regulators of infection [J]. Annu Rev Phytopathol, 2014, 52: 267-288. |
| 84 | Zhang J, Cui WY, Abdul Haseeb H, et al. VdNop12, containing two tandem RNA recognition motif domains, is a crucial factor for pathogenicity and cold adaption in Verticillium dahliae [J]. Environ Microbiol, 2020, 22(12): 5387-5401. |
| 85 | Zhang Y, Liu L, Qiu QZ, et al. Alternative polyadenylation: methods, mechanism, function, and role in cancer [J]. J Exp Clin Cancer Res, 2021, 40(1): 51. |
| 86 | Geng GW, Yu CM, Li XD, et al. Variable 3'polyadenylation of Wheat yellow mosaic virus and its novel effects on translation and replication [J]. Virol J, 2019, 16(1): 23. |
| 87 | Zeng W, Dai XH, Sun J, et al. Modulation of auxin signaling and development by polyadenylation machinery [J]. Plant Physiol, 2019, 179(2): 686-699. |
| 88 | Xu HD, Ning BL, Mu F, et al. Advances of functional consequences and regulation mechanisms of alternative cleavage and polyadenylation [J]. Yi Chuan, 2021, 43(1): 4-15. |
| 89 | Ye CT, Zhou Q, Wu XH, et al. Genome-wide alternative polyadenylation dynamics in response to biotic and abiotic stresses in rice [J]. Ecotoxicol Environ Saf, 2019, 183: 109485. |
| 90 | Yu ZB, Wang J, Zhang C, et al. SIZ1-mediated SUMOylation of CPSF100 promotes plant thermomorphogenesis by controlling alternative polyadenylation [J]. Mol Plant, 2024, 17(9): 1392-1406. |
| 91 | Huang MH, Jiang Y, Qin RF, et al. Full-length transcriptional analysis of the same soybean genotype with compatible and incompatible reactions to Heterodera glycines reveals nematode infection activating plant defense response [J]. Front Plant Sci, 2022, 13: 866322. |
| 92 | Wen JF, Zhu QF, Liu Y, et al. RNA modifications: emerging players in the regulation of reproduction and development [J]. Acta Biochim Biophys Sin, 2024, 57(1): 33-58. |
| 93 | Lee KP, Liu K, Kim EY, et al. The m6A reader ECT1 drives mRNA sequestration to dampen salicylic acid-dependent stress responses in Arabidopsis [J]. Plant Cell, 2024, 36(3): 746-763. |
| 94 | Pessina S, Angeli D, Martens S, et al. The knock-down of the expression of MdMLO19 reduces susceptibility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica) [J]. Plant Biotechnol J, 2016, 14(10): 2033-2044. |
| 95 | Pessina S, Pavan S, Catalano D, et al. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica [J]. BMC Genomics, 2014, 15(1): 618. |
| 96 | Guo TL, Liu CH, Meng FX, et al. The m6A reader MhYTP2 regulates MdMLO19 mRNA stability and antioxidant genes translation efficiency conferring powdery mildew resistance in apple [J]. Plant Biotechnol J, 2022, 20(3): 511-525. |
| 97 | Song ZH, Yang Q, Dong BY, et al. Nanopore RNA direct sequencing identifies that m6A modification is essential for sorbitol-controlled resistance to Alternaria alternata in apple [J]. Dev Cell, 2025. |
| 98 | Zhang TY, Wang ZQ, Hu HC, et al. Transcriptome-wide N6-methyladenosine (m6A) profiling of susceptible and resistant wheat varieties reveals the involvement of variety-specific m6A modification involved in virus-host interaction pathways [J]. Front Microbiol, 2021, 12: 656302. |
| 99 | Zhou LL, Gao GT, Tang RK, et al. m6A-mediated regulation of crop development and stress responses [J]. Plant Biotechnol J, 2022, 20(8): 1447-1455. |
| 100 | Wu HJ, Qu XY, Dong ZC, et al. WUSCHEL triggers innate antiviral immunity in plant stem cells [J]. Science, 2020, 370(6513): 227-231. |
| 101 | Gu HQ, Lian B, Yuan YX, et al. A 5' tRNA-Ala-derived small RNA regulates anti-fungal defense in plants [J]. Sci China Life Sci, 2022, 65(1): 1-15. |
| 102 | Castello A, Fischer B, Frese CK, et al. Comprehensive identification of RNA-binding domains in human cells [J]. Mol Cell, 2016, 63(4): 696-710. |
| 103 | 曾如馨, 陈鹏. RNA结合蛋白的组学解析与功能探索 [J]. 化学学报, 2024, 82(1): 53-61. |
| Zeng RX, Chen P. Genomic analysis and functional exploration of RNA binding protein [J]. China Ind Econ, 2024, 82(1): 53-61. | |
| 104 | Mateos JL, Staiger D. Toward a systems view on RNA-binding proteins and associated RNAs in plants: Guilt by association [J]. Plant Cell, 2023, 35(6): 1708-1726. |
| 105 | Wei CH, Wu MM, Wang CD, et al. Long noncoding RNA lnc-SEMT modulates IGF2 expression by sponging miR-125b to promote sheep muscle development and growth [J]. Cell Physiol Biochem, 2018, 49(2): 447-462. |
| 106 | Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development [J]. Nat Rev Genet, 2014, 15(1): 7-21. |
| 107 | Huang J, Zhou WL, Zhang XM, et al. Roles of long non-coding RNAs in plant immunity [J]. PLoS Pathog, 2023, 19(5): e1011340. |
| 108 | Nejat N, Mantri N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses [J]. Crit Rev Biotechnol, 2018, 38(1): 93-105. |
| 109 | Di C, Yuan JP, Wu Y, et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features [J]. Plant J, 2014, 80(5): 848-861. |
| [1] | ZHANG Na, LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong. Alternative Translation of Wheat Enolase-encoding Gene ENO2 and Its Prokaryotic Expression [J]. Biotechnology Bulletin, 2024, 40(5): 112-119. |
| [2] | GENG Ruo-han, WANG Bing-he, XU Chang-wen, QIAN Hong-ping, LIN Jin-xing, CUI Ya-ning. Research Progress in the Regulation of Protein Post-translational Modification in Plant Vesicle Transport [J]. Biotechnology Bulletin, 2024, 40(10): 139-148. |
| [3] | XUE Jiao ZHU Qing-feng FENG Yan-zhao CHEN Pei LIU Wen-hua ZHANG Ai-xia LIU Qin-jian ZHANG Qi YU Yang. Advances in Upstream Open Reading Frame in Plant Genes [J]. Biotechnology Bulletin, 2023, 39(4): 157-165. |
| [4] | ZHANG Xiao-yan, YANG Shu-hua, DING Yang-lin. Molecular Mechanism of Cold Signal Perception and Transduction in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 28-35. |
| [5] | ZHOU Heng, XIE Yan-jie. Recent Progress in Oxidative Stress Signaling and Response in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 36-43. |
| [6] | JIA Hai-hong, LI Bing-qing. Research Progress in the Post-translational Modification of Superoxide Dismutase [J]. Biotechnology Bulletin, 2022, 38(2): 237-244. |
| [7] | MA Rong, SHANG Fang-zheng, PAN Jian-feng, RONG You-jun, WANG Min, LI Jin-quan, ZHANG Yan-jun. Research Progress in Influencing Factors of mRNA Translation in Cells and Translatome [J]. Biotechnology Bulletin, 2022, 38(12): 115-126. |
| [8] | JIANG Yan-ke, LU Chong-chong, YIN Zi-yi, LI Yang, DING Xin-hua. Research Progress in Alternative Splicing in Plant Immunity [J]. Biotechnology Bulletin, 2022, 38(1): 215-227. |
| [9] | LIU Jing, LI Ya-chao, ZHOU Meng-yan, WU Peng-fei, MA Xiang-qing, LI Ming. Advances in the Studies of Plant Protein Post-translational Modification [J]. Biotechnology Bulletin, 2021, 37(1): 67-76. |
| [10] | ZHANG Cui-ju, MO Bei-xin, CHEN Xue-mei, CUI Jie. Advances on the Molecular Action Mechanisms of Plant miRNA [J]. Biotechnology Bulletin, 2020, 36(7): 1-14. |
| [11] | LI Xiao-yan, LI Chi-yu, YU Feng, LIAO Hong-dong. Preliminary Study on Interaction Between Arabidopsis thaliana EBP1 Protein and RNA [J]. Biotechnology Bulletin, 2020, 36(6): 35-45. |
| [12] | WANG Yong-ping, REN Wei, WANG Run-juan, SHAO Kun-zhong, GAO Hui-juan, ZHANG Jin-lin. Research Advances on Functions of SUMO E3 Ligase in Plant Abiotic Stress Adaptation [J]. Biotechnology Bulletin, 2020, 36(2): 169-177. |
| [13] | BAO Jing-jing, PU Ya-bin, MA Yue-hui, ZHAO Qian-jun. Identification and Analysis of Alternative Splicing in Longissimus dorsi of Sheep at Different Development Stages [J]. Biotechnology Bulletin, 2019, 35(7): 33-38. |
| [14] | LI Zhen-zhen, ZHAO Jia-chen, MA Yu-shu. The Translational Regulation of Gene Expression in Gluconobacter oxydans by Small RNAs [J]. Biotechnology Bulletin, 2018, 34(3): 59-66. |
| [15] | CHEN Jing, ZHANG Dao-wei, QIAN Zheng-min. Structure and Characteristics Analysis of Chitin Synthase 1 from Sogota furcifera [J]. Biotechnology Bulletin, 2018, 34(1): 195-201. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||