








生物技术通报 ›› 2023, Vol. 39 ›› Issue (10): 231-245.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0315
徐俊1( ), 叶雨晴1, 牛雅静2, 黄河1(
), 叶雨晴1, 牛雅静2, 黄河1( ), 张蒙蒙2(
), 张蒙蒙2( )
)
收稿日期:2023-04-08
出版日期:2023-10-26
发布日期:2023-11-28
通讯作者:
黄河,男,博士,教授,研究方向:花卉分子生物学;E-mail: 101navy@163.com;作者简介:徐俊,男,硕士研究生,研究方向:花卉种质资源与遗传育种;E-mail: 1405549321@qq.com
基金资助:
XU Jun1( ), YE Yu-qing1, NIU Ya-jing2, HUANG He1(
), YE Yu-qing1, NIU Ya-jing2, HUANG He1( ), ZHANG Meng-meng2(
), ZHANG Meng-meng2( )
)
Received:2023-04-08
Published:2023-10-26
Online:2023-11-28
摘要:
为了探究菊花根状茎形成的分子机制,本研究选取了具有稳定根状茎的菊花株系‘2017XS’的根状茎尖、根状茎中部、根状茎下部、叶片、茎段、根系、茎尖、舌状花8 个部位进行了转录组测序,并利用生物信息学方法对根状茎尖和整个根状茎中高表达的基因进行筛选。转录组测序共得到159.51 GB 数据,组装后得到100 235个Unigene。其中,64 956(64.80%)个Unigene在7个公共蛋白质数据库中得到了注释。为了找到根状茎发育的关键基因,通过加权基因共表达网络分析(weighted correlation network analysis,WGCNA)、K-means和基于Venn筛选差异基因3种方式筛选在根状茎尖和根状茎中高表达基因。最终筛选到20个在根状茎尖中高表达的基因和36个在根状茎中高表达的基因。这些基因包括了和植物非生物胁迫相关的基因、脱落酸(abscisic acid, ABA)代谢基因、红光受体和紫外光受体以及光周期核心转录因子,选取6个差异表达较显著的基因进行 RT-qPCR 荧光定量分析,结果与转录组测序数据一致,验证了转录组的有效性。且这些差异表达基因在同样具根茎的菊花株系‘2005042’中亦表现为在根状茎中特异高表达。综上,菊花根状茎的形成和发育可能受到包括脱落酸在内的植物激素以及光周期等因素的影响,本研究对进一步探究菊花根状茎发育的分子机制提供了重要依据。
徐俊, 叶雨晴, 牛雅静, 黄河, 张蒙蒙. 菊花根状茎发育的转录组分析[J]. 生物技术通报, 2023, 39(10): 231-245.
XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium[J]. Biotechnology Bulletin, 2023, 39(10): 231-245.
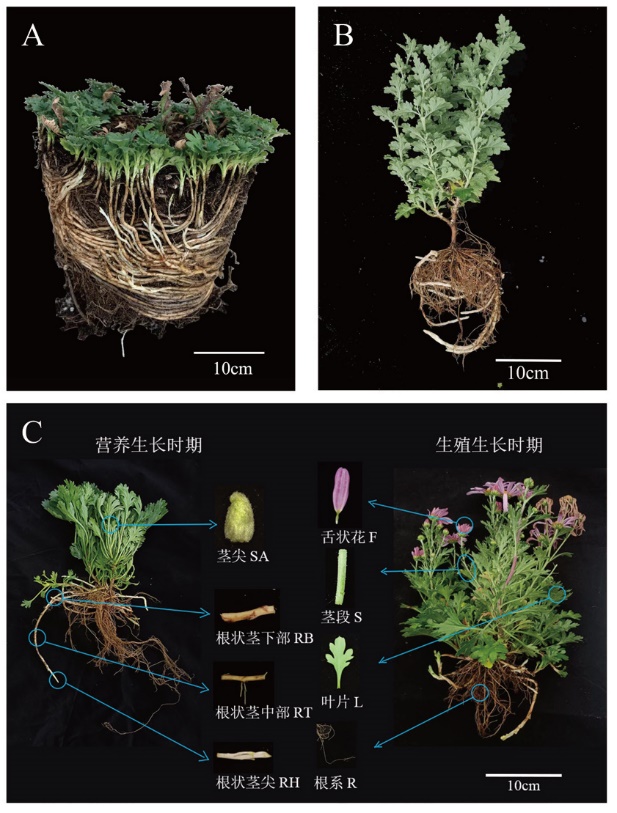
图1 ‘2017XS’和‘2005042’根状茎示意图及转录组取材示意图 A:根状茎株系‘2017XS’;B:根状茎株系‘2005042’;C:用于转录组测序的材料为‘2017XS’株系。蓝圈表示取材的具体部位,箭头指向该部位放大后的细部图。除舌状花外其他材料均取自营养生长时期
Fig. 1 Schematic diagram of ‘2017XS’ and ‘2005042’ rhizomes and schematic diagram of 8 transcriptome sampling A: Strain with rhizome‘2017XS’. B: Strain with rhizome‘2005042’. C: The material used for transcriptome sequencing is‘2017XS’. The blue circle indicates the specific part of the material, and the arrow indicates the details of the material. Ray florets were taken from the reproductive stage, and other materials were taken from the vegetative stage
| 基因名称Gene name | 基因序列号Gene ID | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|---|
| LHY | c226991.graph_c0 | CGAAGACCGCAGTGCAAATC | TCCCATCCTTTTCTGCCACC |
| BG | c188673.graph_c0 | TGGTCTCGGTTGTTTTGCTTG | TGGAAATGAAGACGGAGTGGAA |
| bZIP | c208538.graph_c0 | GCAATACATATCGGAGCTAGAACG | GCAATCCTTTGCTTGAGGACAC |
| ABA8ox | c209725.graph_c0 | AAGCTCGTTCAAGGCTCGTTAT | TGAGTCCAAGGCGGAGACAG |
| UGT | c177448.graph_c0 | TAACAAGTATGGAAGGAGCAGGTG | GTGGATGGGATGGATGACGA |
| RLP | c217842.graph_c0 | GGATTTCGGGAATGCACTTACT | TCTAATGGAATCGGACCTGCTAAT |
表1 RT-qPCR引物序列
Table 1 Primer sequences for RT-qPCR
| 基因名称Gene name | 基因序列号Gene ID | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|---|
| LHY | c226991.graph_c0 | CGAAGACCGCAGTGCAAATC | TCCCATCCTTTTCTGCCACC |
| BG | c188673.graph_c0 | TGGTCTCGGTTGTTTTGCTTG | TGGAAATGAAGACGGAGTGGAA |
| bZIP | c208538.graph_c0 | GCAATACATATCGGAGCTAGAACG | GCAATCCTTTGCTTGAGGACAC |
| ABA8ox | c209725.graph_c0 | AAGCTCGTTCAAGGCTCGTTAT | TGAGTCCAAGGCGGAGACAG |
| UGT | c177448.graph_c0 | TAACAAGTATGGAAGGAGCAGGTG | GTGGATGGGATGGATGACGA |
| RLP | c217842.graph_c0 | GGATTTCGGGAATGCACTTACT | TCTAATGGAATCGGACCTGCTAAT |
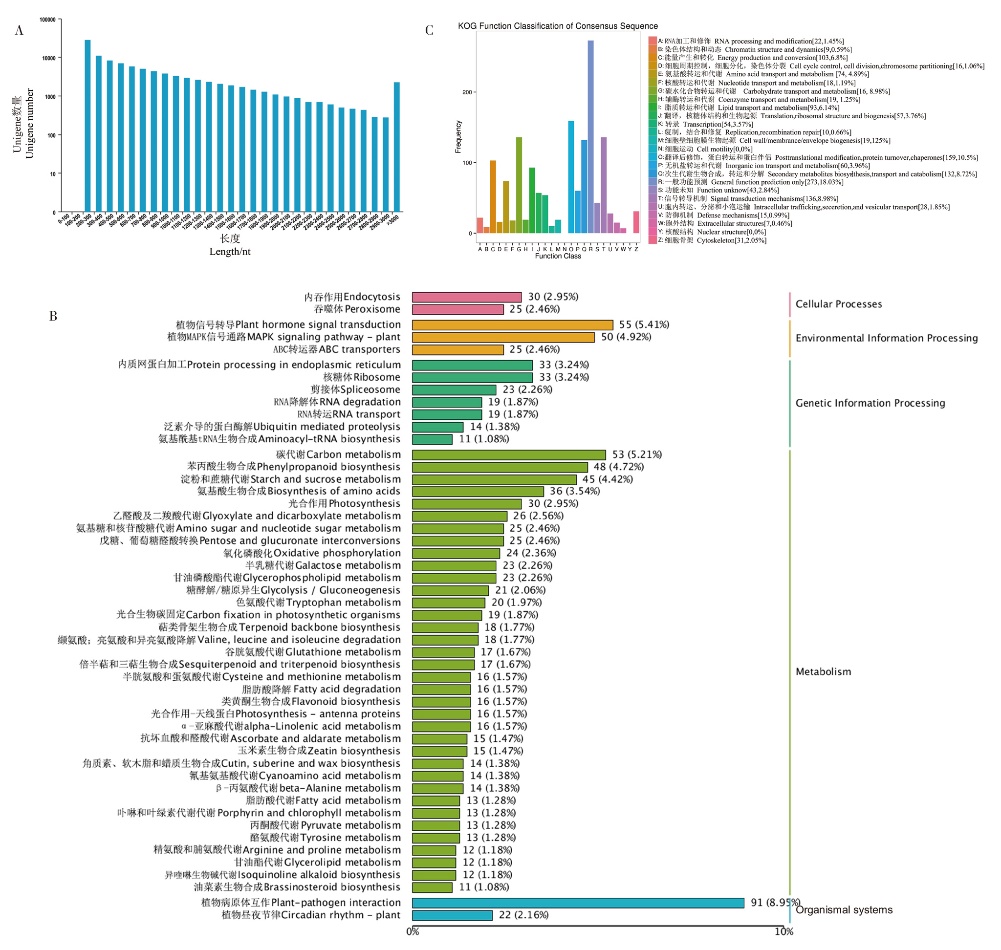
图2 所有Unigene长度分布及根状茎差异基因的功能注释和分类 A:转录组中Unigene在不同长度的分布情况。横坐标表示Unigene的长度,纵坐标表示不同长度Unigene的数量;B:根状茎差异基因的KOG分析直方图。匹配到KOG数据库的Unigne被分到25个类别。不同颜色的柱状图表示不同的KOG类别,纵坐标表示该类别中基因出现的频数;C:根状茎差异基因的KEGG通路分类。不同的柱状图表示不同KEGG通路分类,粉色柱状图表示细胞过程,黄色柱状图表示环境信息过程,深绿色柱状图表示遗传信息过程,浅绿色柱状图表示代谢过程,蓝色柱状图表示有机体系统。数字分别表示该KEGG类别上的基因数量和该类别基因数量在所有基因数量中的百分比
Fig. 2 Unigene length distribution and functional annotation and classification of DEGs in rhizomes A: The distribution of Unigene in different lengths in transcriptome. The X-axis represents the length of Unigene, and the Y-axis represents the number of Unigenes at different lengths. B: Histogram of KOG classification. Unigenes with significant matches in the KOG database are classified into 25 categories. Histograms of different colors represent different KOG categories, and Y-axis represent the number of genes. C: Classification of Unigenes in KEGG pathways. The pink histogram represents the Cellular Process, the yellow histogram represents the Environmental Information Process, the dark green histogram represents the Genetic Information Process, the light green histogram represents the Metabolism, and the blue histogram represents the Organismal system. The number represents the number of genes in this KEGG category and the percentage of the number of genes in this category in the total number of genes
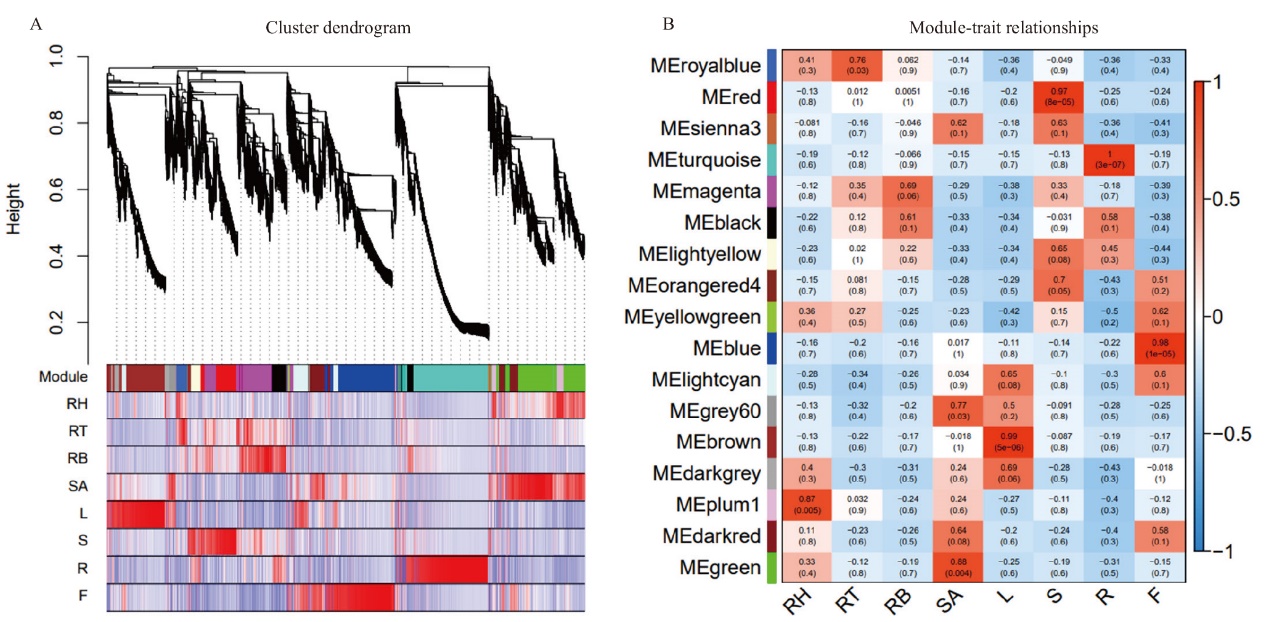
图3 加权基因共表达网络分析 A:系统聚类树和模块划分。上部为基因聚类树,下部是按树的分枝切割得到的模块,相同的模块用同一种颜色表示;B:基因共表达模块与2017XS各组织间的相关性。每行代表一个模块,每列代表一个组织,矩形框里的数字代表模块与性状之间的相关系数及相应P值,红色表示模块与组织正相关性,蓝色表示负相关。RH:根状茎尖;RT:根状茎中部;RB:根状茎下部;SA:茎尖;L:叶片;S:茎段;R:根系;F:舌状花
Fig. 3 WGCNA analysis A: Clustering dendrograms of genes and module division. The upper part is the genes cluster dendrograms, the lower part is assigned module, and the same modules had the same color. B: Heat map of the correlation between co-expressed modules and‘2017XS’ tissues. Each row represents a module, and each column represents a tissue. The number in the rectangular box represents the correlation coefficient and corresponding P-value between the module and the trait. Red block represents the positive correlation between the module and the tissues, and blue represents the negative correlation. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret
| 差异基因筛选方式 Screening method of DEGs | 基因 Gene | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | PME68(c195055.graph_c0) | Probable pectinesterase 68(Artemisia annua) | 23.06 | 4.01 | 0.19 | 6.81 | 0.12 | 0.35 | 3.85 | 0.06 |
| PHYB(c200625.graph_c0) | Phytochrome B(Artemisia annua) | 5.83 | 1.45 | 0.57 | 1.18 | 0 | 0.58 | 0 | 0.85 | |
| FLA7(c216061.graph_c0) | Fasciclin-like arabinogalactan protein 7(Tanacetum cinerariifolium) | 71.37 | 7.08 | 0.08 | 14.25 | 4.05 | 9.19 | 0.02 | 19.08 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| OLE(c190721.graph_c0) | Oleosin(Artemisia annua) | 5.17 | 1.52 | 0.24 | 0.7 | 0.04 | 0.21 | 0 | 0.81 | |
| TIP2-1(c156293.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 24.91 | 2.7 | 0.29 | 0 | 0 | 0.32 | 0 | 0.32 | |
| LIL3.1(c197767.graph_c1) | Light-harvesting complex-like protein 3 isotype 1(Helianthus annuus) | 7.43 | 0.18 | 0.15 | 0.17 | 0.22 | 0.15 | 0.82 | 0.03 | |
| ABA8ox(c209725.graph_c0) | ABA 8-oxidase(Artemisia annua) | 12.55 | 1.35 | 0.27 | 0.68 | 0.39 | 2.05 | 0.1 | 1.04 | |
| TIP1-1(c185069.graph_c0) | Aquaporin TIP1-1(Artemisia annua) | 503.78 | 74.87 | 6.23 | 51.4 | 32.75 | 21.61 | 0.04 | 6.6 | |
| RD21A(c208052.graph_c0) | Cysteine proteinase COT44(Tanacetum cinerariifolium) | 13.76 | 1.99 | 1.56 | 0.98 | 0.28 | 1.71 | 0.12 | 0.37 | |
| ARP1(c197835.graph_c0) | Probable RNA-binding protein ARP1 isoform X2(Tanacetum cinerariifolium) | 10.34 | 0.74 | 0.07 | 1.05 | 0 | 0.95 | 0 | 0.13 | |
| ARP1(c197835.graph_c1) | Probable RNA-binding protein ARP1 isoform X2(Helianthus annuus) | 50.65 | 6.38 | 0.65 | 4.54 | 0.05 | 5.96 | 0.1 | 0.95 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| bZIP34(c208538.graph_c0) | Basic leucine zipper 34(Artemisia annua) | 41.51 | 11.57 | 0.27 | 2.55 | 2.05 | 7.91 | 0 | 2.58 | |
| HIDH(c211020.graph_c0) | 2-hydroxyisoflavanone dehydratase-like(Erigeron canadensis) | 7.48 | 1.18 | 1.16 | 1.96 | 0.08 | 1.97 | 2.16 | 0.03 | |
| VENN | DREB2F(c193648.graph_c0) | Dehydration-responsive element-binding protein 2F(Tanacetum cinerariifolium) | 3.13 | 0.03 | 0.03 | 0.38 | 0 | 0 | 0 | 0 |
| CZF2(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.3 | 0.3 | 0.24 | 0.06 | 0.2 | |
| ABA8ox4(c212882.graph_c0) | Abscisic acid 8’-hydroxylase 4(Tanacetum cinerariifolium) | 3.72 | 0.63 | 0.07 | 0.11 | 0.7 | 0.72 | 0.12 | 0.06 | |
| PP2C38(c204872.graph_c0) | Probable protein phosphatase 2C 38(Artemisia annua) | 5.27 | 0.08 | 0.02 | 0.16 | 0.7 | 0 | 0.02 | 0.16 | |
| UGT83A1(c177448.graph_c0) | UDP-glucosyltransferase(Artemisia annua) | 2.8 | 0.06 | 0.1 | 0.25 | 0.23 | 0.05 | 0 | 0 | |
表2 ‘2017XS’根状茎尖中高表达基因汇总
Table 2 Summary of highly expressed genes in the rhizome apical of 2017XS
| 差异基因筛选方式 Screening method of DEGs | 基因 Gene | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | PME68(c195055.graph_c0) | Probable pectinesterase 68(Artemisia annua) | 23.06 | 4.01 | 0.19 | 6.81 | 0.12 | 0.35 | 3.85 | 0.06 |
| PHYB(c200625.graph_c0) | Phytochrome B(Artemisia annua) | 5.83 | 1.45 | 0.57 | 1.18 | 0 | 0.58 | 0 | 0.85 | |
| FLA7(c216061.graph_c0) | Fasciclin-like arabinogalactan protein 7(Tanacetum cinerariifolium) | 71.37 | 7.08 | 0.08 | 14.25 | 4.05 | 9.19 | 0.02 | 19.08 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| OLE(c190721.graph_c0) | Oleosin(Artemisia annua) | 5.17 | 1.52 | 0.24 | 0.7 | 0.04 | 0.21 | 0 | 0.81 | |
| TIP2-1(c156293.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 24.91 | 2.7 | 0.29 | 0 | 0 | 0.32 | 0 | 0.32 | |
| LIL3.1(c197767.graph_c1) | Light-harvesting complex-like protein 3 isotype 1(Helianthus annuus) | 7.43 | 0.18 | 0.15 | 0.17 | 0.22 | 0.15 | 0.82 | 0.03 | |
| ABA8ox(c209725.graph_c0) | ABA 8-oxidase(Artemisia annua) | 12.55 | 1.35 | 0.27 | 0.68 | 0.39 | 2.05 | 0.1 | 1.04 | |
| TIP1-1(c185069.graph_c0) | Aquaporin TIP1-1(Artemisia annua) | 503.78 | 74.87 | 6.23 | 51.4 | 32.75 | 21.61 | 0.04 | 6.6 | |
| RD21A(c208052.graph_c0) | Cysteine proteinase COT44(Tanacetum cinerariifolium) | 13.76 | 1.99 | 1.56 | 0.98 | 0.28 | 1.71 | 0.12 | 0.37 | |
| ARP1(c197835.graph_c0) | Probable RNA-binding protein ARP1 isoform X2(Tanacetum cinerariifolium) | 10.34 | 0.74 | 0.07 | 1.05 | 0 | 0.95 | 0 | 0.13 | |
| ARP1(c197835.graph_c1) | Probable RNA-binding protein ARP1 isoform X2(Helianthus annuus) | 50.65 | 6.38 | 0.65 | 4.54 | 0.05 | 5.96 | 0.1 | 0.95 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| bZIP34(c208538.graph_c0) | Basic leucine zipper 34(Artemisia annua) | 41.51 | 11.57 | 0.27 | 2.55 | 2.05 | 7.91 | 0 | 2.58 | |
| HIDH(c211020.graph_c0) | 2-hydroxyisoflavanone dehydratase-like(Erigeron canadensis) | 7.48 | 1.18 | 1.16 | 1.96 | 0.08 | 1.97 | 2.16 | 0.03 | |
| VENN | DREB2F(c193648.graph_c0) | Dehydration-responsive element-binding protein 2F(Tanacetum cinerariifolium) | 3.13 | 0.03 | 0.03 | 0.38 | 0 | 0 | 0 | 0 |
| CZF2(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.3 | 0.3 | 0.24 | 0.06 | 0.2 | |
| ABA8ox4(c212882.graph_c0) | Abscisic acid 8’-hydroxylase 4(Tanacetum cinerariifolium) | 3.72 | 0.63 | 0.07 | 0.11 | 0.7 | 0.72 | 0.12 | 0.06 | |
| PP2C38(c204872.graph_c0) | Probable protein phosphatase 2C 38(Artemisia annua) | 5.27 | 0.08 | 0.02 | 0.16 | 0.7 | 0 | 0.02 | 0.16 | |
| UGT83A1(c177448.graph_c0) | UDP-glucosyltransferase(Artemisia annua) | 2.8 | 0.06 | 0.1 | 0.25 | 0.23 | 0.05 | 0 | 0 | |
| 差异基因 筛选方式 Screening method of DEGs | 基因序列号 Gene ID | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | TFL1(c181499.graph_c0) | TFL1-like protein(Chrysanthemum x morifolium) | 6.19 | 14.30 | 1.24 | 0.14 | 0.20 | 0.07 | 0.17 | 0.04 |
| UVR8(c225307.graph_c0) | Ultraviolet-B receptor UVR8 isoform X2(Cynara cardunculus) | 24.60 | 21.64 | 27.53 | 1.41 | 1.62 | 1.30 | 0.74 | 1.52 | |
| GWD(c220138.graph_c0) | Alpha-glucan water dikinase 1(Helianthus annuus) | 31.29 | 22.66 | 22.05 | 7.28 | 5.81 | 0.10 | 4.84 | 4.22 | |
| UGT73C3(c193961.graph_c0) | UDP-glucosyl transferase 73B2(Lactuca sativa) | 11.45 | 24.51 | 12.62 | 1.14 | 0.49 | 0.65 | 2.62 | 0.32 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| LEA Dc3(c195245.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 3668.15 | 3341 | 1742.08 | 13.18 | 8.53 | 2.06 | 16.2 | 9.52 | |
| zinc finger(c171393.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 1.82 | 15.18 | 3.16 | 0 | 0 | 0.29 | 0.99 | 0.51 | |
| RLP7(c217842.graph_c0) | Leucine-rich repeat protein(Artemisia annua) | 14.87 | 9.42 | 2.48 | 0.02 | 0.07 | 0.09 | 0.13 | 0 | |
| ERF098(c180295.graph_c0) | Ethylene-responsive transcription factor ERF098(Lactuca sativa) | 3.51 | 3.40 | 2.77 | 0.88 | 0.41 | 0 | 0.48 | 0 | |
| PIP1-1(c215772.graph_c0) | Aquaporin PIP1-1-like(Artemisia annua) | 343.98 | 533.32 | 370.29 | 44.36 | 10.71 | 48.83 | 15.59 | 32.56 | |
| hAT transposon(c228735.graph_c0) | Putative hAT transposon superfamily(Tanacetum cinerariifolium) | 895.49 | 1500.66 | 580.71 | 95.56 | 57.82 | 32.38 | 158.83 | 76.66 | |
| GH1(c176119.graph_c0) | Glycoside hydrolase family 1(Artemisia annua) | 12.04 | 27.68 | 15.34 | 0 | 0.13 | 1.02 | 3.03 | 0.73 | |
| SAMDC(c206787.graph_c0) | S-adenosylmethionine decarboxylase proenzyme(Artemisia annua) | 88.53 | 53.73 | 22.24 | 2.34 | 1.76 | 1.26 | 2.14 | 1.16 | |
| LNK1(c223971.graph_c0) | Protein LNK1-like isoform X1(Lactuca sativa) | 14.82 | 14.45 | 16.06 | 1.13 | 0.59 | 1.93 | 0.32 | 0.41 | |
| LHY(c226991.graph_c0) | Late elongated hypocotyl-like(Chrysanthemum seticuspe f. boreale) | 55.07 | 59.39 | 47.02 | 11.46 | 6.01 | 8.76 | 4.67 | 12.26 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| RE1(c213815.graph_c1) | Putative RNA-directed DNA polymerase(Helianthus annuus) | 14.31 | 9.52 | 8.36 | 1.19 | 0.32 | 1.76 | 1.61 | 0.38 | |
| BG 46(c188673.graph_c0) | Beta glucosidase 46(Artemisia annua) | 5.00 | 7.08 | 7.03 | 0.15 | 0.80 | 0 | 0.11 | 0 | |
| ACA8(c226840.graph_c0) | Calcium-transporPting ATPase 8(Erigeron canadensis) | 65.4 | 106.47 | 58.25 | 12.76 | 6.78 | 8.67 | 14.69 | 16.91 | |
| DREB1D(c212875.graph_c0) | Dehydration-responsive element-binding factor 1(Artemisia annua) | 13.93 | 8.66 | 2.94 | 0.16 | 0 | 0.04 | 0.25 | 0.13 | |
| MLP43(c91334.graph_c0) | MLP-like protein 43(Tanacetum cinerariifolium) | 128.75 | 232.78 | 41.83 | 8.21 | 0 | 1.35 | 9.98 | 3.82 | |
| K-means | LEA14(c204348.graph_c0) | Late embryogenesis abundant protein(Artemisia annua) | 972.25 | 411.65 | 189.58 | 2.30 | 4.40 | 0.19 | 12.78 | 5.00 |
| ACA10(c210148.graph_c0) | Calcium-transporting ATPase 10(Tanacetum cinerariifolium) | 60.68 | 126.95 | 65.40 | 13.2 | 4.11 | 8.93 | 16.34 | 19.69 | |
| LYK4(c215208.graph_c1) | LysM domain receptor-like kinase 4(Tanacetum cinerariifolium) | 26.77 | 49.34 | 43.47 | 1.80 | 1.66 | 6.83 | 7.13 | 0.44 | |
| PDC2(c187399.graph_c0) | Pyruvate decarboxylase 2(Tanacetum cinerariifolium) | 1.05 | 8.54 | 5.68 | 0 | 0.12 | 0 | 0 | 0 | |
| UGT73(c215576.graph_c0) | UDP-glycosyltransferase 73C7-like(Cynara cardunculus) | 29.51 | 59.45 | 37.95 | 3.22 | 2.8 | 2.83 | 5.55 | 1.07 | |
| zinc finger(c193154.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 13.11 | 17.47 | 14.45 | 2.30 | 0.10 | 0.29 | 3.16 | 1.62 | |
| UVR8(c192386.graph_c0) | Ultraviolet-B receptor UVR8-like isoform X1(Erigeron canadensis) | 19.07 | 21.94 | 29.46 | 0.41 | 0.45 | 0.63 | 0 | 0 | |
| 1-SST(c222090.graph_c0) | Sucrose:sucrose 1-fructosyl transferase(Artemisia annua) | 1005.96 | 2853.86 | 1238.04 | 1.80 | 4.62 | 16.83 | 79.72 | 297.35 | |
| MLP43(c195694.graph_c0) | MLP-like protein 43(Artemisia annua) | 2035.38 | 5638.39 | 3866.43 | 224.77 | 17.43 | 77.31 | 669.73 | 75.88 | |
| COL12(c204664.graph_c0) | B-box zinc finger protein 32(Artemisia annua) | 2.50 | 3.98 | 4.90 | 0.14 | 0 | 0 | 0.08 | 0 | |
| VENN | CZF4(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.30 | 0.30 | 0.06 | 0.24 | 0.20 |
| ERF C3(c195922.graph_c0) | Ethylene-response factor C3(Helianthus annuus) | 1.13 | 4.14 | 8.48 | 0 | 0 | 0.27 | 0.20 | 0.04 | |
| NDA1(c208704.graph_c0) | NADH dehydrogenase(Artemisia annua) | 1.63 | 1.43 | 3.08 | 0.21 | 0.06 | 0 | 0.46 | 0.27 | |
| BG 46(c196303.graph_c0) | Putative beta-glucosidase(Helianthus annuus) | 1.54 | 3.26 | 1.97 | 0.06 | 0.06 | 0.18 | 0.39 | 0.03 | |
| LEA Dc3(c208154.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 0.19 | 2.36 | 51.39 | 0.05 | 0 | 0.04 | 0 | 0 | |
表3 ‘2017XS’根状茎中高表达基因汇总
Table 3 Summary of highly expressed genes in the rhizomes of ‘2017XS’
| 差异基因 筛选方式 Screening method of DEGs | 基因序列号 Gene ID | 功能注释 Functional annotation | FPKM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 根状茎尖 Rhizome tip | 根状茎中部 Rhizome middle | 根状茎下部 Rhizome bottom | 茎尖 Shoot apical | 叶片 Leaf | 茎段 Stem | 根系 Root | 舌状花 Ray floret | |||
| WGCNA | TFL1(c181499.graph_c0) | TFL1-like protein(Chrysanthemum x morifolium) | 6.19 | 14.30 | 1.24 | 0.14 | 0.20 | 0.07 | 0.17 | 0.04 |
| UVR8(c225307.graph_c0) | Ultraviolet-B receptor UVR8 isoform X2(Cynara cardunculus) | 24.60 | 21.64 | 27.53 | 1.41 | 1.62 | 1.30 | 0.74 | 1.52 | |
| GWD(c220138.graph_c0) | Alpha-glucan water dikinase 1(Helianthus annuus) | 31.29 | 22.66 | 22.05 | 7.28 | 5.81 | 0.10 | 4.84 | 4.22 | |
| UGT73C3(c193961.graph_c0) | UDP-glucosyl transferase 73B2(Lactuca sativa) | 11.45 | 24.51 | 12.62 | 1.14 | 0.49 | 0.65 | 2.62 | 0.32 | |
| LEA1(c165828.graph_c1) | Late embryogenesis abundant protein 1(Artemisia annua) | 295.49 | 72.99 | 9.25 | 0.34 | 0.73 | 0 | 0 | 0 | |
| LEA Dc3(c195245.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 3668.15 | 3341 | 1742.08 | 13.18 | 8.53 | 2.06 | 16.2 | 9.52 | |
| zinc finger(c171393.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 1.82 | 15.18 | 3.16 | 0 | 0 | 0.29 | 0.99 | 0.51 | |
| RLP7(c217842.graph_c0) | Leucine-rich repeat protein(Artemisia annua) | 14.87 | 9.42 | 2.48 | 0.02 | 0.07 | 0.09 | 0.13 | 0 | |
| ERF098(c180295.graph_c0) | Ethylene-responsive transcription factor ERF098(Lactuca sativa) | 3.51 | 3.40 | 2.77 | 0.88 | 0.41 | 0 | 0.48 | 0 | |
| PIP1-1(c215772.graph_c0) | Aquaporin PIP1-1-like(Artemisia annua) | 343.98 | 533.32 | 370.29 | 44.36 | 10.71 | 48.83 | 15.59 | 32.56 | |
| hAT transposon(c228735.graph_c0) | Putative hAT transposon superfamily(Tanacetum cinerariifolium) | 895.49 | 1500.66 | 580.71 | 95.56 | 57.82 | 32.38 | 158.83 | 76.66 | |
| GH1(c176119.graph_c0) | Glycoside hydrolase family 1(Artemisia annua) | 12.04 | 27.68 | 15.34 | 0 | 0.13 | 1.02 | 3.03 | 0.73 | |
| SAMDC(c206787.graph_c0) | S-adenosylmethionine decarboxylase proenzyme(Artemisia annua) | 88.53 | 53.73 | 22.24 | 2.34 | 1.76 | 1.26 | 2.14 | 1.16 | |
| LNK1(c223971.graph_c0) | Protein LNK1-like isoform X1(Lactuca sativa) | 14.82 | 14.45 | 16.06 | 1.13 | 0.59 | 1.93 | 0.32 | 0.41 | |
| LHY(c226991.graph_c0) | Late elongated hypocotyl-like(Chrysanthemum seticuspe f. boreale) | 55.07 | 59.39 | 47.02 | 11.46 | 6.01 | 8.76 | 4.67 | 12.26 | |
| TIP2-1(c181494.graph_c0) | Aquaporin TIP2-1(Tanacetum cinerariifolium) | 13.52 | 0.95 | 0.16 | 0 | 0 | 0 | 0 | 0 | |
| RE1(c213815.graph_c1) | Putative RNA-directed DNA polymerase(Helianthus annuus) | 14.31 | 9.52 | 8.36 | 1.19 | 0.32 | 1.76 | 1.61 | 0.38 | |
| BG 46(c188673.graph_c0) | Beta glucosidase 46(Artemisia annua) | 5.00 | 7.08 | 7.03 | 0.15 | 0.80 | 0 | 0.11 | 0 | |
| ACA8(c226840.graph_c0) | Calcium-transporPting ATPase 8(Erigeron canadensis) | 65.4 | 106.47 | 58.25 | 12.76 | 6.78 | 8.67 | 14.69 | 16.91 | |
| DREB1D(c212875.graph_c0) | Dehydration-responsive element-binding factor 1(Artemisia annua) | 13.93 | 8.66 | 2.94 | 0.16 | 0 | 0.04 | 0.25 | 0.13 | |
| MLP43(c91334.graph_c0) | MLP-like protein 43(Tanacetum cinerariifolium) | 128.75 | 232.78 | 41.83 | 8.21 | 0 | 1.35 | 9.98 | 3.82 | |
| K-means | LEA14(c204348.graph_c0) | Late embryogenesis abundant protein(Artemisia annua) | 972.25 | 411.65 | 189.58 | 2.30 | 4.40 | 0.19 | 12.78 | 5.00 |
| ACA10(c210148.graph_c0) | Calcium-transporting ATPase 10(Tanacetum cinerariifolium) | 60.68 | 126.95 | 65.40 | 13.2 | 4.11 | 8.93 | 16.34 | 19.69 | |
| LYK4(c215208.graph_c1) | LysM domain receptor-like kinase 4(Tanacetum cinerariifolium) | 26.77 | 49.34 | 43.47 | 1.80 | 1.66 | 6.83 | 7.13 | 0.44 | |
| PDC2(c187399.graph_c0) | Pyruvate decarboxylase 2(Tanacetum cinerariifolium) | 1.05 | 8.54 | 5.68 | 0 | 0.12 | 0 | 0 | 0 | |
| UGT73(c215576.graph_c0) | UDP-glycosyltransferase 73C7-like(Cynara cardunculus) | 29.51 | 59.45 | 37.95 | 3.22 | 2.8 | 2.83 | 5.55 | 1.07 | |
| zinc finger(c193154.graph_c0) | Zinc finger, CCHC-type(Artemisia annua) | 13.11 | 17.47 | 14.45 | 2.30 | 0.10 | 0.29 | 3.16 | 1.62 | |
| UVR8(c192386.graph_c0) | Ultraviolet-B receptor UVR8-like isoform X1(Erigeron canadensis) | 19.07 | 21.94 | 29.46 | 0.41 | 0.45 | 0.63 | 0 | 0 | |
| 1-SST(c222090.graph_c0) | Sucrose:sucrose 1-fructosyl transferase(Artemisia annua) | 1005.96 | 2853.86 | 1238.04 | 1.80 | 4.62 | 16.83 | 79.72 | 297.35 | |
| MLP43(c195694.graph_c0) | MLP-like protein 43(Artemisia annua) | 2035.38 | 5638.39 | 3866.43 | 224.77 | 17.43 | 77.31 | 669.73 | 75.88 | |
| COL12(c204664.graph_c0) | B-box zinc finger protein 32(Artemisia annua) | 2.50 | 3.98 | 4.90 | 0.14 | 0 | 0 | 0.08 | 0 | |
| VENN | CZF4(c195263.graph_c0) | Zinc finger, CCCH-type(Artemisia annua) | 4.12 | 0.99 | 0.95 | 0.30 | 0.30 | 0.06 | 0.24 | 0.20 |
| ERF C3(c195922.graph_c0) | Ethylene-response factor C3(Helianthus annuus) | 1.13 | 4.14 | 8.48 | 0 | 0 | 0.27 | 0.20 | 0.04 | |
| NDA1(c208704.graph_c0) | NADH dehydrogenase(Artemisia annua) | 1.63 | 1.43 | 3.08 | 0.21 | 0.06 | 0 | 0.46 | 0.27 | |
| BG 46(c196303.graph_c0) | Putative beta-glucosidase(Helianthus annuus) | 1.54 | 3.26 | 1.97 | 0.06 | 0.06 | 0.18 | 0.39 | 0.03 | |
| LEA Dc3(c208154.graph_c0) | Late embryogenesis abundant protein Dc3(Artemisia annua) | 0.19 | 2.36 | 51.39 | 0.05 | 0 | 0.04 | 0 | 0 | |

图4 根状茎3个部位与其他组织的差异表达基因Venn图 A:根状茎尖和非根状茎部位差异基因;B:根状茎中部和非根状茎部位差异基因;C:根状茎下部和非根状茎部位差异基因。不同颜色的集合表示不同的比较组,数字表示该区域内包含的基因数量。红色方框表示在五个比较组中都差异表达的基因。RH:根状茎上部;RT:根状茎中部;RB:根状茎下部;SA:茎尖; L:叶片;S:茎段;R:根系;F:舌状花
Fig. 4 Venn diagram of DEGs in three parts of rhizome and other tissues A: DEGs between rhizome tip and non-rhizome tissues. B: DEGs between middle of rhizome and non-rhizome tissues. C: DEGs between bottle of rhizome and non-rhizome tissues. Sets of different colors represent different comparison groups, and numbers represent the number of genes contained in the region. The red box indicates the genes differentially expressed in the five comparison groups. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret

图5 根状茎稳定型株系‘2017XS’基因表达模式的Cluster分析 每个聚类的横坐标表示转录组的取材测序组织,纵坐标表示该聚类内基因的表达量,曲线表示该聚类内基因表达量的变化趋势。RH:根状茎上部;RT:根状茎中部;RB:根状茎下部;SA:茎尖;L:叶片;S:茎段;R:根系;F:舌状花
Fig. 5 Cluster analysis of gene expression patterns of rhizome stable line ‘2017XS’ The X-axis of each cluster represents the material sequencing organization of the transcriptome, the Y-axis represents the gene expression level in the cluster, and the curve represents the change trend of gene expression level in the cluster. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret
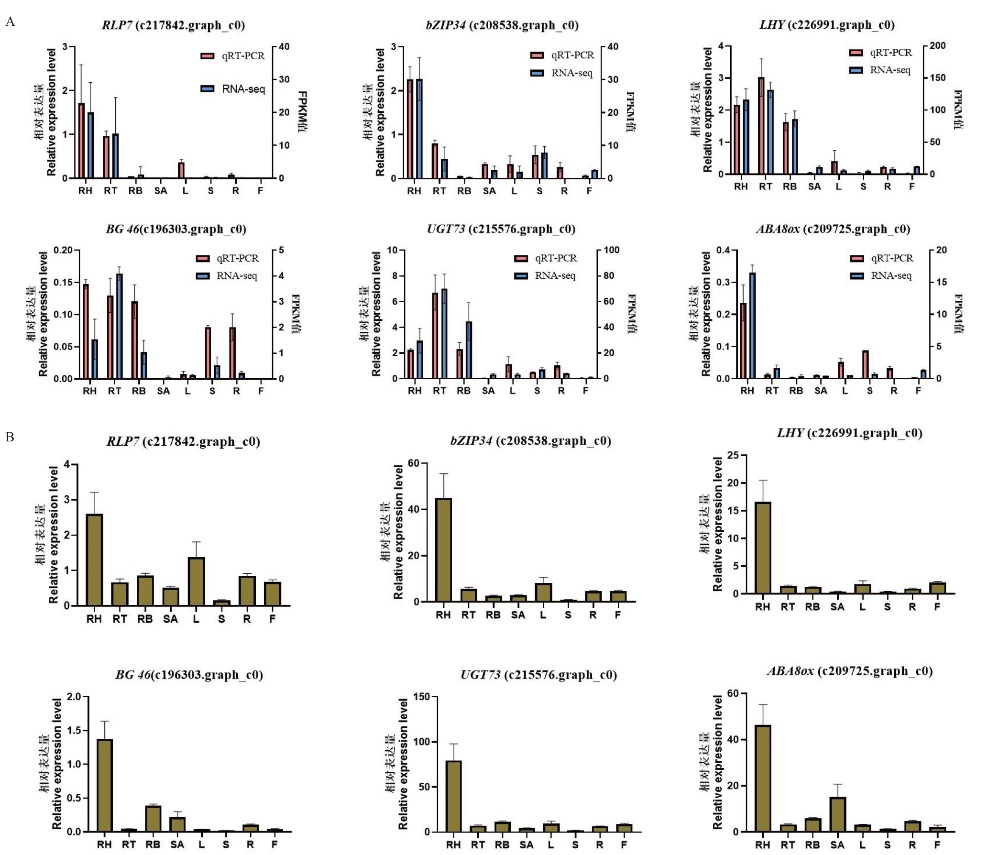
图6 转录组测序数据表达验证 A:转录组筛选基因在‘2017XS’中的RT-qPCR表达验证;B:转录组筛选基因在‘2005042’中的RT-qPCR表达验证。RH:根状茎上部;RT:根状茎中部;RB:根状茎下部;SA:茎尖;L:叶片;S:茎段;R:根系;F:舌状花
Fig. 6 Verification of transcriptome sequencing data A: RT-qPCR verification of transcriptome-screened genes in ‘2017XS’. B: RT-qPCR verification of transcriptome-screened genes in‘2005042’. RH: rhizome apical; RT: middle of rhizome; RB: bottle of rhizome; SA: shoot apical; L: leaf; S: shoot; R: root; F: ray floret
| [1] |
Yang W, Liu WH, Niu KJ, et al. Transcriptional regulation of different rhizome parts reveal the candidate genes that regulate rhizome development in Poa pratensis[J]. DNA Cell Biol, 2022, 41(2): 151-168.
doi: 10.1089/dna.2021.0337 URL |
| [2] |
Yoshida A, Terada Y, Toriba T, et al. Analysis of rhizome development in Oryza longistaminata, a wild rice species[J]. Plant Cell Physiol, 2016, 57(10): 2213-2220.
pmid: 27516415 |
| [3] |
Ma XQ, Yu JJ, Zhuang LL, et al. Differential regulatory pathways associated with drought-inhibition and post-drought recuperation of rhizome development in perennial grass[J]. Ann Bot, 2020, 126(3): 481-497.
doi: 10.1093/aob/mcaa099 URL |
| [4] |
Zhang SL, Huang GF, Zhang YJ, et al. Sustained productivity and agronomic potential of perennial rice[J]. Nat Sustain, 2023, 6(1): 28-38.
doi: 10.1038/s41893-022-00997-3 |
| [5] |
Anderson N, Gesick E. Phenotypic markers for selection of winter hardy garden chrysanthemum(Dendranthema × grandiflora Tzvelv.) genotypes[J]. Sci Hortic, 2004, 101(1/2): 153-167.
doi: 10.1016/j.scienta.2003.10.006 URL |
| [6] |
Zhang LL, Xu YJ, Liu XN, et al. The chrysanthemum DEAD-box RNA helicase CmRH56 regulates rhizome outgrowth in response to drought stress[J]. J Exp Bot, 2022, 73(16): 5671-5681.
doi: 10.1093/jxb/erac213 URL |
| [7] |
Fu JX, Yang LW, Dai SL. Conservation of Arabidopsis thaliana circadian clock genes in Chrysanthemum lavandulifolium[J]. Plant Physiol Biochem, 2014, 80: 337-347.
doi: 10.1016/j.plaphy.2014.04.001 URL |
| [8] |
Fu JX, Yang LW, Dai SL. Identification and characterization of the CONSTANS-like gene family in the short-day plant Chrysanthemum lavandulifolium[J]. Mol Genet Genomics, 2015, 290(3): 1039-1054.
doi: 10.1007/s00438-014-0977-3 URL |
| [9] |
Hong Y, Yang LW, Li ML, et al. Comparative analyses of light-induced anthocyanin accumulation and gene expression between the ray florets and leaves in chrysanthemum[J]. Plant Physiol Biochem, 2016, 103: 120-132.
doi: 10.1016/j.plaphy.2016.03.006 URL |
| [10] |
Yang M, Zhu LP, Pan C, et al. Transcriptomic analysis of the regulation of rhizome formation in temperate and tropical lotus(Nelumbo nucifera)[J]. Sci Rep, 2015, 5: 13059.
doi: 10.1038/srep13059 pmid: 26279185 |
| [11] |
Cheng LB, Li SY, Yin JJ, et al. Genome-wide analysis of differentially expressed genes relevant to rhizome formation in lotus root(Nelumbo nucifera gaertn)[J]. PLoS One, 2013, 8(6): e67116.
doi: 10.1371/journal.pone.0067116 URL |
| [12] |
Zhang T, Zhao XQ, Wang WS, et al. Deep transcriptome sequencing of rhizome and aerial-shoot in Sorghum propinquum[J]. Plant Mol Biol, 2014, 84(3): 315-327.
doi: 10.1007/s11103-013-0135-z pmid: 24104862 |
| [13] |
Wang KH, Peng HZ, Lin EP, et al. Identification of genes related to the development of bamboo rhizome bud[J]. J Exp Bot, 2010, 61(2): 551-561.
doi: 10.1093/jxb/erp334 pmid: 19965904 |
| [14] |
Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nat Biotechnol, 2011, 29(7): 644-652.
doi: 10.1038/nbt.1883 pmid: 21572440 |
| [15] |
Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Res, 1997, 25(17): 3389-3402.
doi: 10.1093/nar/25.17.3389 pmid: 9254694 |
| [16] |
Xie C, Mao XZ, Huang JJ, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases[J]. Nucleic Acids Res, 2011, 39(Web Server issue): W316-W322.
doi: 10.1093/nar/gkr483 URL |
| [17] |
Eddy SR. HMMER: profile HMMs for protein sequence analysis. Bioinformatics, 1998, 14: 755-763.
doi: 10.1093/bioinformatics/14.9.755 pmid: 9918945 |
| [18] |
Yu GC, Wang LG, Han YY, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16(5): 284-287.
doi: 10.1089/omi.2011.0118 pmid: 22455463 |
| [19] |
Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome[J]. Genome Biol, 2009, 10(3): R25.
doi: 10.1186/gb-2009-10-3-r25 URL |
| [20] |
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12: 323.
doi: 10.1186/1471-2105-12-323 pmid: 21816040 |
| [41] |
Osnato M, Cota I, Nebhnani P, et al. Photoperiod control of plant growth: flowering time genes beyond flowering[J]. Front Plant Sci, 2022, 12: 805635.
doi: 10.3389/fpls.2021.805635 URL |
| [42] |
Andrés J, Caruana J, Liang JH, et al. Woodland strawberry axillary bud fate is dictated by a crosstalk of environmental and endogenous factors[J]. Plant Physiol, 2021, 187(3): 1221-1234.
doi: 10.1093/plphys/kiab421 pmid: 34618090 |
| [43] |
Zhou TT, Song BT, Liu TF, et al. Phytochrome F plays critical roles in potato photoperiodic tuberization[J]. Plant J, 2019, 98(1): 42-54.
doi: 10.1111/tpj.14198 |
| [21] |
Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data[J]. Bioinformation, 2007, 2(1): 5-7.
doi: 10.6026/97320630002005 pmid: 18084642 |
| [22] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] |
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9: 559.
doi: 10.1186/1471-2105-9-559 pmid: 19114008 |
| [24] | Hu RB, Yu CJ, Wang XY, et al. De novo transcriptome analysis of Miscanthus lutarioriparius identifies candidate genes in rhizome development[J]. Front Plant Sci, 2017, 8: 492. |
| [25] |
Huang H, Wang Y, Wang SL, et al. Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium[J]. Plant Sci, 2012, 193/194: 18-27.
doi: 10.1016/j.plantsci.2012.05.004 URL |
| [26] |
Chai Q, Jin F, Merewitz E, et al. Growth and physiological traits associated with drought survival and post-drought recovery in perennial turfgrass species[J]. J Amer Soc Hort Sci, 2010, 135(2): 125-133.
doi: 10.21273/JASHS.135.2.125 URL |
| [27] | 张黎黎. 菊花DEAD-box RNA解旋酶基因CmRH56调节地下茎发生与干旱耐性的功能分析[D]. 北京: 中国农业大学, 2017. |
| Zhang LL. Function analysis of DEAD-box RNA helicase CmRH56 regulating morphogenesis of suckers and drought stress tolerance in Chrysanthemum[D]. Beijing: China Agricultural University, 2017. | |
| [28] |
Zhou YB, Chen M, Guo JK, et al. Overexpression of soybean DREB1 enhances drought stress tolerance of transgenic wheat in the field[J]. J Exp Bot, 2020, 71(6): 1842-1857.
doi: 10.1093/jxb/erz569 URL |
| [29] |
Wang GD, Xu XP, Wang H, et al. A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato[J]. Plant Physiol Biochem, 2019, 142: 254-262.
doi: 10.1016/j.plaphy.2019.07.017 URL |
| [30] |
Lim J, Lim CW, Lee SC. The pepper late embryogenesis abundant protein, CaDIL1, positively regulates drought tolerance and ABA signaling[J]. Front Plant Sci, 2018, 9: 1301.
doi: 10.3389/fpls.2018.01301 pmid: 30233631 |
| [31] | 袁娅娟. 草地早熟禾根茎扩展与内源激素及碳氮代谢随生育时期的动态变化[D]. 兰州: 甘肃农业大学, 2021. |
| Yuan YJ. Study on the dynamic changes of rhizome expansion, endogenous hormones, carbon and nitrogen metabolism of Poa pratensis during growth stage[D]. Lanzhou: Gansu Agricultural University, 2021. | |
| [32] |
Li LS, Xia TZ, Li B, et al. Hormone and carbohydrate metabolism associated genes play important roles in rhizome bud full-year germination of Cephalostachyum pingbianense[J]. Physiol Plant, 2022, 174(2): e13674.
doi: 10.1111/ppl.v174.2 URL |
| [33] |
Ma XQ, Huang BR. Gibberellin-stimulation of rhizome elongation and differential GA-responsive proteomic changes in two grass species[J]. Front Plant Sci, 2016, 7: 905.
doi: 10.3389/fpls.2016.00905 pmid: 27446135 |
| [34] |
Zdunek-Zastocka E, Grabowska A. The interplay of PsABAUGT1 with other abscisic acid metabolic genes in the regulation of ABA homeostasis during the development of pea seeds and germination in the presence of H2O2[J]. Plant Sci, 2019, 285: 79-90.
doi: S0168-9452(19)30178-5 pmid: 31203896 |
| [35] |
Zheng CL, Acheampong AK, Shi ZW, et al. Abscisic acid catabolism enhances dormancy release of grapevine buds[J]. Plant Cell Environ, 2018, 41(10): 2490-2503.
doi: 10.1111/pce.v41.10 URL |
| [36] | Wang J, Xu YD, Yin ZN, et al. Overexpression of the persimmon abscisic acid DkUGT3 gene alters plant/fruit development in transgenic tomato[J]. J Plant Growth Regul, 2023: 1-15. |
| [37] |
Liang B, Zheng Y, Wang J, et al. Overexpression of the persimmon abscisic acid β-glucosidase gene(DkBG1)alters fruit ripening in transgenic tomato[J]. Plant J, 2020, 102(6): 1220-1233.
doi: 10.1111/tpj.v102.6 URL |
| [38] |
Masuda JI, Ozaki Y, Okubo H. Rhizome transition to storage organ is under phytochrome control in lotus(Nelumbo nucifera)[J]. Planta, 2007, 226(4): 909-915.
doi: 10.1007/s00425-007-0536-9 URL |
| [39] |
Saxena P, Huang BR, Bonos SA, et al. Photoperiod and temperature effects on rhizome production and tillering rate in tall fescue[Lolium arundinaceum(schreb.) darby.[J]. Crop Sci, 2014, 54(3): 1205-1210.
doi: 10.2135/cropsci2013.08.0565 URL |
| [40] |
Xu ZE, Chen HJ, Ji LF, et al. Polymorphisms of the FT gene as a tool to identify underground rhizome types of bamboos[J]. Euphytica, 2017, 213(1): 25.
doi: 10.1007/s10681-016-1824-x |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [3] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [4] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [5] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [6] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [7] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [8] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [9] | 孙言秋, 谢采芸, 汤岳琴. 耐高温酿酒酵母的构建与高温耐受机制解析[J]. 生物技术通报, 2023, 39(11): 226-237. |
| [10] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [11] | 辛建攀, 李燕, 赵楚, 田如男. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210. |
| [12] | 许瑾, 李涛, 李楚琳, 朱顺妮, 王忠铭, 向文洲. 温度对真眼点藻生长、总脂及二十碳五烯酸(EPA)合成的影响[J]. 生物技术通报, 2022, 38(6): 261-271. |
| [13] | 熊和丽, 沙茜, 刘韶娜, 相德才, 张斌, 赵智勇. 单细胞转录组测序技术在动物上的应用研究[J]. 生物技术通报, 2022, 38(3): 226-233. |
| [14] | 张斌, 杨昕霞. 水稻响应盐胁迫关键转录因子的鉴定[J]. 生物技术通报, 2022, 38(3): 9-15. |
| [15] | 关怡, 李新, 王定一, 杜茜, 张龙斌, 叶秀云. BbRho5对球孢白僵菌生长速率的作用研究[J]. 生物技术通报, 2022, 38(2): 132-140. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||