








生物技术通报 ›› 2022, Vol. 38 ›› Issue (6): 129-135.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0963
收稿日期:2021-07-28
出版日期:2022-06-26
发布日期:2022-07-11
基金资助:Received:2021-07-28
Published:2022-06-26
Online:2022-07-11
摘要:
探讨拟南芥GRAS转录因子AtSCL4(Arabidopsis thaliana SCL4)在渗透胁迫中发挥的生物学功能,为GRAS蛋白在非生物胁迫中的功能研究奠定基础。以野生型和AtSCL4突变体拟南芥为试验材料,通过生理指标测定和qRT-PCR方法研究渗透胁迫下AtSCL4调控植物抗逆的生物学机制。研究发现AtSCL4受渗透胁迫诱导后显著上调表达,且AtSCL4突变体的抗渗能力强于野生型。在渗透胁迫下,AtSCL4可以负调节ATMYB6的表达,减小气孔开放度,降低叶片水分流失;AtSCL4通过负调控P5SC1和BADH的转录来提高植物体内脯氨酸和甜菜碱的含量;AtSCL4通过负调节AtSOD1和PER4的表达来增加抗氧化酶活性而降低活性氧含量。AtSCL4可负向调控抗逆基因表达和生理变化应答渗透胁迫。
徐红云, 张明意. GRAS转录因子AtSCL4负调控拟南芥应答渗透胁迫[J]. 生物技术通报, 2022, 38(6): 129-135.
XU Hong-yun, ZHANG Ming-yi. AtSCL4,an Arabidopsis thaliana GRAS Transcription Factor,Negatively Modulates Plants in Response to Osmotic Stress[J]. Biotechnology Bulletin, 2022, 38(6): 129-135.
| 引物名称 Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| AtSCL4-F | ATGGCTTACATGTGCACTGATAG |
| AtSCL4-R | TTATCGCCAGGAAGAAAGAGTG |
| ATMYB61-F | GATTGCGTCAAGGCTTCCG |
| ATMYB61-R | CGCAGAAGAGGAACTAGGAG |
| P5CS1-F | GTCAAGTCTATGCTTGATTTG |
| P5CS1-R | GATTTGTCGCCGAATGTAATC |
| BADH-F | GCTGACCTAGCTGAAGGCTTG |
| BADH-R | CACCTCGCGGCAAATATCAGC |
| SOD1-F | GTCCACATTTCAACCCCGATG |
| SOD1-R | GAGACCAATGATGCCGCAAGC |
| PER4-F | GAAGGTTGGTCGAAGAGATTC |
| PER4-R | CGTATCTCCACCGTTGACCGG |
| Act7-F | ATGTTCACCACTACCGCAG |
| Act7-R | ACCTCAGGACAACGGAATCTC |
| Tub2-F | GTCTCCAAGGGTTCCAGGTT |
| Tub2-R | GACAGAGTAGCGTTGTAAGGC |
表1 qRT-PCR所用引物序列
Table 1 Primer sequences used in qRT-PCR
| 引物名称 Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| AtSCL4-F | ATGGCTTACATGTGCACTGATAG |
| AtSCL4-R | TTATCGCCAGGAAGAAAGAGTG |
| ATMYB61-F | GATTGCGTCAAGGCTTCCG |
| ATMYB61-R | CGCAGAAGAGGAACTAGGAG |
| P5CS1-F | GTCAAGTCTATGCTTGATTTG |
| P5CS1-R | GATTTGTCGCCGAATGTAATC |
| BADH-F | GCTGACCTAGCTGAAGGCTTG |
| BADH-R | CACCTCGCGGCAAATATCAGC |
| SOD1-F | GTCCACATTTCAACCCCGATG |
| SOD1-R | GAGACCAATGATGCCGCAAGC |
| PER4-F | GAAGGTTGGTCGAAGAGATTC |
| PER4-R | CGTATCTCCACCGTTGACCGG |
| Act7-F | ATGTTCACCACTACCGCAG |
| Act7-R | ACCTCAGGACAACGGAATCTC |
| Tub2-F | GTCTCCAAGGGTTCCAGGTT |
| Tub2-R | GACAGAGTAGCGTTGTAAGGC |
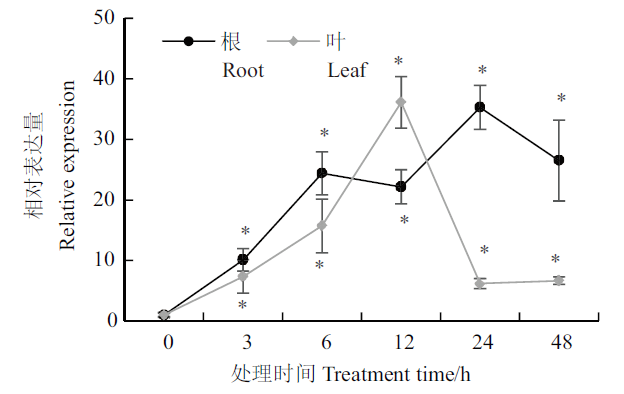
图1 甘露醇处理下AtSCL4基因在不同时间段的表达分析 *:在P<0.05水平差异显著。下同
Fig. 1 Expression profiles of AtSCL4 at different times in response to mannitol stress *:Indicates difference significant at P< 0.05. The same below
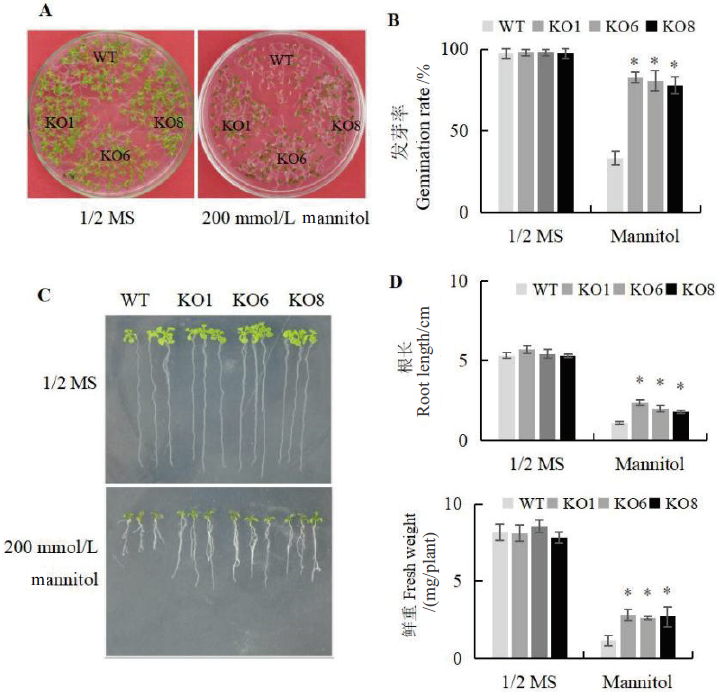
图3 渗透胁迫下AtSCL4调控苗的萌发和生长分析 A:平板培养基观察萌发率情况;B:发芽率统计;C:根长观察;D:根长和鲜重统计
Fig. 3 Analysis of seedling germination and growth regulated by AtSCL4 under osmotic stress A:Observation of the germination phenotypes on plate medium. B:Statistics of germination. C:Observation of root length. D:Statistics of root length and fresh weight
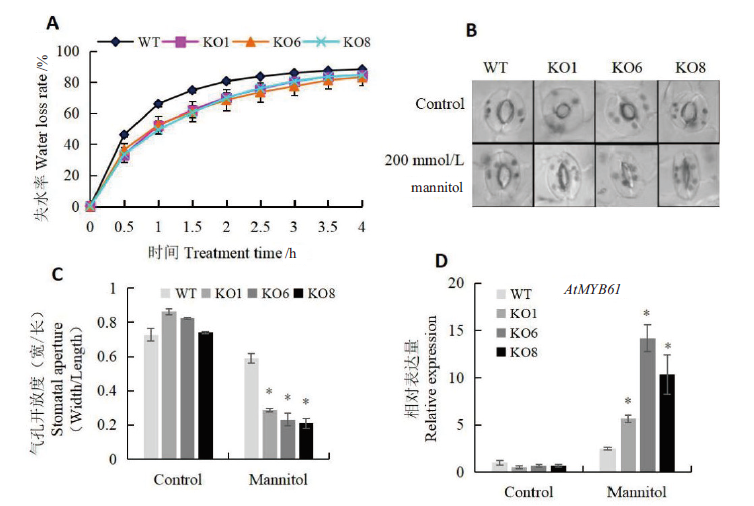
图4 渗透胁迫下AtSCL4调控苗的气孔开放度分析 A:叶片失水率测定;B:气孔观察;C:气孔开放度统计;D:气孔调控相关基因AtMYB61表达量分析
Fig. 4 Stomatal aperture assay of seedlings regulated by AtSCL4 under osmotic stress A:Determination of water loss rate. B:Observation of stomatal aperture.C:Determination of stomatal aperture. D:Expression of AtMYB61 related to stomatal aperture regulation
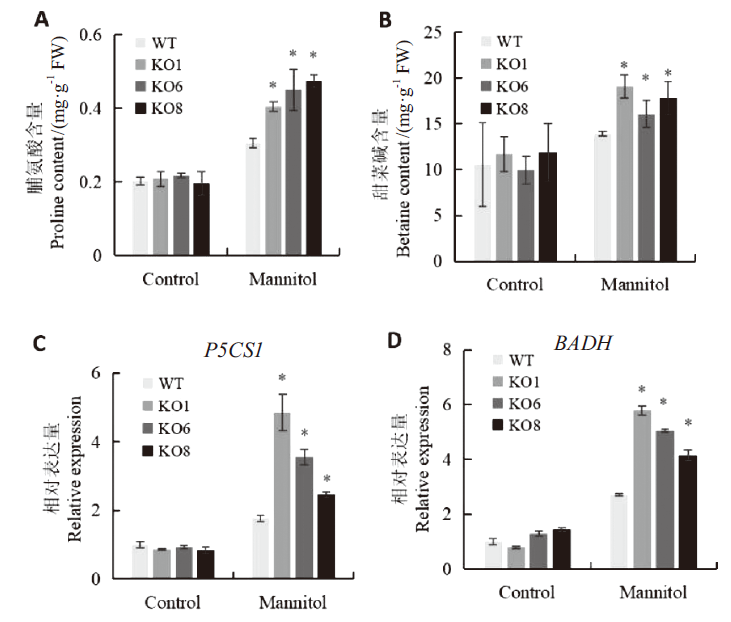
图5 渗透胁迫下AtSCL4调控苗渗透物质合成分析 A:脯氨酸含量;B:甜菜碱含量;C:脯氨酸合成相关基因P5CS1表达量分析;D:甜菜碱合成相关基因BADH表达量分析
Fig. 5 Synthesis analysis of osmolyte in seedlings mediated by AtSCL4 under osmotic stress A:Proline content. B:Betaine content. C:Expressions of of P5CS1 related to proline synthesis. D:Expressions of BADH related to betaine synthesis
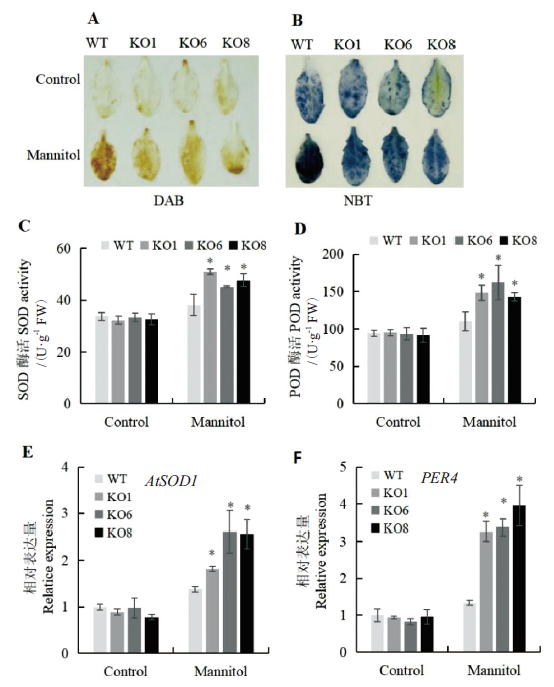
图6 渗透胁迫下AtSCL4调控苗活性氧清除能力分析 A:DAB染色观察H2O2含量;B:NBT染色观察O2-·含量;C:SOD酶活;D:POD酶活;E:SOD酶合成相关基因AtSOD1表达量分析;F:POD酶合成相关基因PER4表达量分析
Fig. 6 ROS scavenging capacity assay of seedlings regul-ated by AtSCL4 under osmotic stress A:Observation of H2O2 content by DAB staining. B:Observation of O2-· content by NBT staining. C:SOD activities. D:POD activities. E:Expression of AtSOD1.F:Expression of PER4
| [1] |
Wang J, Song L, Gong X, et al. Functions of jasmonic acid in plant regulation and response to abiotic stress[J]. Int J Mol Sci, 2020, 21(4):1446.
doi: 10.3390/ijms21041446 URL |
| [2] |
Rabbani MA, Maruyama K, Abe H, et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses[J]. Plant Physiol, 2003, 133(4):1755-1767.
pmid: 14645724 |
| [3] |
Hsieh EJ, Cheng MC, Lin TP. Functional characterization of an abiotic stress-inducible transcription factor AtERF53 in Arabidopsis thaliana[J]. Plant Mol Biol, 2013, 82(3):223-237.
doi: 10.1007/s11103-013-0054-z URL |
| [4] |
Sakuraba Y, Kim YS, Han SH, et al. The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP[J]. Plant Cell, 2015, 27(6):1771-1787.
doi: 10.1105/tpc.15.00222 URL |
| [5] |
Bolle C. The role of GRAS proteins in plant signal transduction and development[J]. Planta, 2004, 218(5):683-692.
doi: 10.1007/s00425-004-1203-z URL |
| [6] |
Sun X, Xue B, Jones WT, et al. A functionally required unfoldome from the plant kingdom:intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development[J]. Plant Mol Biol, 2011, 77(3):205-223.
doi: 10.1007/s11103-011-9803-z URL |
| [7] |
Liu X, Widmer A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and rice[J]. Plant Mol Biol Report, 2014, 32(6):1129-1145.
doi: 10.1007/s11105-014-0721-5 URL |
| [8] |
Chen YQ, Tai SS, Wang DW, et al. Homology-based analysis of the GRAS gene family in tobacco[J]. Genet Mol Res, 2015, 14(4):15188-15200.
doi: 10.4238/2015.November.25.7 pmid: 26634482 |
| [9] |
Lu J, Wang T, Xu Z, et al. Genome-wide analysis of the GRAS gene family in Prunus mume[J]. Mol Genet Genomics, 2015, 290(1):303-317.
doi: 10.1007/s00438-014-0918-1 URL |
| [10] |
Song XM, Liu TK, Duan WK, et al. Genome-wide analysis of the GRAS gene family in Chinese cabbage(Brassica rapa ssp. pekinensis)[J]. Genomics, 2014, 103(1):135-146.
doi: 10.1016/j.ygeno.2013.12.004 URL |
| [11] |
Huang W, Xian Z, Kang X, et al. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato[J]. BMC Plant Biol, 2015, 15:209.
doi: 10.1186/s12870-015-0590-6 pmid: 26302743 |
| [12] | Wang Y, Shi S, Zhou Y, et al. Genome-wide identification and characterization of GRAS transcription factors in sacred Lotus(Nelumbo nucifera)[J]. PeerJ, 2016, 4:e2388. |
| [13] |
Xu K, Chen SJ, Li TF, et al. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes[J]. BMC Plant Biol, 2015, 15:141.
doi: 10.1186/s12870-015-0532-3 URL |
| [14] |
Yuan YY, Fang LC, Karungo SK, et al. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis[J]. Plant Cell Rep, 2016, 35(3):655-666.
doi: 10.1007/s00299-015-1910-x URL |
| [15] |
Ma HS, Liang D, Shuai P, et al. The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana[J]. J Exp Bot, 2010, 61(14):4011-4019.
doi: 10.1093/jxb/erq217 URL |
| [16] |
Zhang S, Li XW, de Fan S, et al. Overexpression of HcSCL13, a Halostachys caspica GRAS transcription factor, enhances plant growth and salt stress tolerance in transgenic Arabidopsis[J]. Plant Physiol Biochem, 2020, 151:243-254.
doi: 10.1016/j.plaphy.2020.03.020 URL |
| [17] | Zhang Y, Zhou YY, Zhang D, et al. PtrWRKY75 overexpression reduces stomatal aperture and improves drought tolerance by salicylic acid-induced reactive oxygen species accumulation in poplar[J]. Environ Exp Bot, 2020, 176:104117. |
| [18] |
Liang YK, Dubos C, Dodd IC, et al. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana[J]. Curr Biol, 2005, 15(13):1201-1206.
doi: 10.1016/j.cub.2005.06.041 URL |
| [19] |
Miller G, Suzuki N, Ciftci-Yilmaz S, et al. Reactive oxygen species homeostasis and signalling during drought and salinity stresses[J]. Plant Cell Environ, 2010, 33(4):453-467.
doi: 10.1111/j.1365-3040.2009.02041.x URL |
| [20] |
Ashraf M, Foolad MR. Roles of Glycine betaine and proline in improving plant abiotic stress resistance[J]. Environ Exp Bot, 2007, 59(2):206-216.
doi: 10.1016/j.envexpbot.2005.12.006 URL |
| [21] |
Silva-Ortega CO, Ochoa-Alfaro AE, Reyes-Agüero JA, et al. Salt stress increases the expression of p5cs gene and induces proline accumulation in Cactus pear[J]. Plant Physiol Biochem, 2008, 46(1):82-92.
doi: 10.1016/j.plaphy.2007.10.011 URL |
| [22] | Ishitani M, Arakawa K, Mizuno K, et al. Betaine aldehyde dehydrogenase in the Gramineae:levels in leaves both betaine-accumulating and nonaccumulating cereal plants[J]. Plant Cell Physiol, 1993, 34(3):493-495. |
| [23] | Petrov V, Hille J, Mueller-Roeber B, et al. ROS-mediated abiotic stress-induced programmed cell death in plants[J]. Front Plant Sci, 2015, 6:69. |
| [24] |
Wang L, Li Z, Lu M, et al. ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis[J]. Front Plant Sci, 2017, 8:635.
doi: 10.3389/fpls.2017.00635 URL |
| [1] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [2] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [3] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [4] | 撒世娟, 伍涵宇, 温媛, 陈雪娜, 郑蕊, 姚新灵. 叶绿体特异蛋白质表达谱对本氏烟不同气孔密度的响应[J]. 生物技术通报, 2023, 39(2): 193-202. |
| [5] | 周恒, 谢彦杰. 植物氧化胁迫信号应答的研究进展[J]. 生物技术通报, 2023, 39(11): 36-43. |
| [6] | 赵佳, 赵飞燕, 沈馨, 高广琦, 孙志宏. 乳酸菌抗氧化活性及其应用研究进展[J]. 生物技术通报, 2023, 39(11): 182-190. |
| [7] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [8] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [9] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [10] | 高聪, 萧楚健, 鲁帅, 王苏蓉, 袁卉华, 曹云英. 氧化石墨烯对拟南芥生长的促进作用[J]. 生物技术通报, 2022, 38(6): 120-128. |
| [11] | 古盼, 齐学影, 李莉, 张曦, 单晓昳. AtRGS1胞吞动态调控G蛋白参与拟南芥生长发育和抗性反应[J]. 生物技术通报, 2022, 38(6): 34-42. |
| [12] | 周娟, 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英. 拟南芥F-box蛋白FKF1与转录因子FUL互作调控开花研究[J]. 生物技术通报, 2022, 38(3): 1-8. |
| [13] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [14] | 李兵娟, 郑璐, 沈仁芳, 兰平. 拟南芥RPP1A参与幼苗生长的蛋白质组学分析[J]. 生物技术通报, 2022, 38(2): 10-20. |
| [15] | 徐子涵, 刘倩, 苗大鹏, 陈跃, 胡凤荣. 春兰miR396过表达对拟南芥叶片生长、光合及叶绿素荧光特性的影响[J]. 生物技术通报, 2021, 37(5): 28-37. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
