








生物技术通报 ›› 2023, Vol. 39 ›› Issue (7): 131-142.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0015
王帅1( ), 冯宇梅2, 白苗1, 杜维俊1, 岳爱琴1(
), 冯宇梅2, 白苗1, 杜维俊1, 岳爱琴1( )
)
收稿日期:2023-01-09
出版日期:2023-07-26
发布日期:2023-08-17
通讯作者:
岳爱琴,女,博士,教授,研究方向:大豆遗传与种质创新;E-mail: yueaiqinnd@126.com作者简介:王帅,男,硕士研究生,研究方向:大豆遗传与种质创新;E-mail: 15234570975@163.com
基金资助:
WANG Shuai1( ), FENG Yu-mei2, BAI Miao1, DU Wei-jun1, YUE Ai-qin1(
), FENG Yu-mei2, BAI Miao1, DU Wei-jun1, YUE Ai-qin1( )
)
Received:2023-01-09
Published:2023-07-26
Online:2023-08-17
摘要:
GmHMGR基因是大豆甲羟戊酸途径的关键酶基因之一,在大豆逆境胁迫调控方面具有重要的作用。探究GmHMGR基因在逆境胁迫中的功能,为大豆抗氧化、抗盐机制解析和分子育种提供依据。采用实时荧光定量PCR、酵母转化、拟南芥遗传转化等技术,分析GmHMGR基因在响应外源激素和非生物胁迫中发挥的功能。结果显示,GmHMGR基因对PEG6000、NaCl、H2O2胁迫以及MeJA、ABA外源激素处理均有响应;过量表达GmHMGR1、GmHMGR3、GmHMGR5、GmHMGR6和GmHMGR7可以提高酵母的抗盐、抗氧化能力;过量表达GmHMGR4与GmHMGR6的拟南芥上调了萜类代谢相关酶基因(AtDXR、AtSQS1、AtSQS2与AtCAS)的表达水平,改善了拟南芥在氧化及盐胁迫下的表型。GmHMGR基因可能参与大豆抗氧化、抗盐胁迫的应答。
王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142.
WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses[J]. Biotechnology Bulletin, 2023, 39(7): 131-142.
| 基因Gene | 登录号Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| GmActin | XM_003552652 | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA |
| qPCR-GmHMGR1 | XM_003517069.4 | GAAAAATCTTACTGGGTCTGCTATG | CTGATTGAGACGCCAGTTGTG |
| qPCR-GmHMGR2 | XM_014775260.2 | TGCTGGTTCTGCTGTCGC | CAAGTAAATTCAAGCAAGCAGATTA |
| qPCR-GmHMGR3 | XM_003537651.3 | TTTCTCCAGAGTGATTTTCC | CAGACCCAGTAAGGTTTTTC |
| qPCR-GmHMGR4 | XM_003534178.4 | TTCTTCAGAGTGACTTCC | GTCTTCAACACCTTCTTC |
| qPCR-GmHMGR5 | XM_006605513.3 | TTCTTCGGCATCCACTTCGTCCAG | GCTGTGCGTCTCCTTCTATGATGG |
| qPCR-GmHMGR6 | XM_003547838.4 | AAGCCGCAGCCGTGAATTGG | CCACCAAGAGCACCAGCCATG |
| qPCR-GmHMGR7 | XM_003519426.4 | TTCGGACAAGAAACCTGC | AAACCCACCAAGAGCACC |
| qPCR-GmHMGR8 | XM_003545508.4 | GCCGTTGTTGTTGGATGG | GCAATCCCTCAAAACCACA |
| AtActin | XM_021028231.1 | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| qPCR-AtDXS | NM_117647.3 | GAAGTCGCAAAGGGTATGACAAAGCA | CTGGATCAAATTTCACAACACCATGGTAT |
| qPCR-AtDXR | NM_125674.3 | TTCTGCCATATTTCAGTGTATTCAAGGTT | GCACAGATGAATCCTGTGTTTCAATC |
| qPCR-AtSQS1 | NM_119630.4 | GGAGAAGCAGATCCCTCCTG | AGTAGAACACACACGGCG |
| qPCR-AtSQS2 | NM_119631.3 | ATCTCAAAGCAAACCAATGTAAG | AGAACAACTGAAAAGGAAGAGAG |
| qPCR-AtCAS | NM_126681.3 | GACGGAGGTTGGGGTTTA | TGATTTAGTATCCAGTCTCGTCC |
| qPCR-AtIPI | NM_121649.6 | TGGGATCATGTTGAGAAAGGAAC | GTTGCCCAGTTTTGTCTGTAATCA |
表 1 实时荧光定量引物序列
Table 1 Primers sequences by real-time quantitative PCR
| 基因Gene | 登录号Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|---|
| GmActin | XM_003552652 | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA |
| qPCR-GmHMGR1 | XM_003517069.4 | GAAAAATCTTACTGGGTCTGCTATG | CTGATTGAGACGCCAGTTGTG |
| qPCR-GmHMGR2 | XM_014775260.2 | TGCTGGTTCTGCTGTCGC | CAAGTAAATTCAAGCAAGCAGATTA |
| qPCR-GmHMGR3 | XM_003537651.3 | TTTCTCCAGAGTGATTTTCC | CAGACCCAGTAAGGTTTTTC |
| qPCR-GmHMGR4 | XM_003534178.4 | TTCTTCAGAGTGACTTCC | GTCTTCAACACCTTCTTC |
| qPCR-GmHMGR5 | XM_006605513.3 | TTCTTCGGCATCCACTTCGTCCAG | GCTGTGCGTCTCCTTCTATGATGG |
| qPCR-GmHMGR6 | XM_003547838.4 | AAGCCGCAGCCGTGAATTGG | CCACCAAGAGCACCAGCCATG |
| qPCR-GmHMGR7 | XM_003519426.4 | TTCGGACAAGAAACCTGC | AAACCCACCAAGAGCACC |
| qPCR-GmHMGR8 | XM_003545508.4 | GCCGTTGTTGTTGGATGG | GCAATCCCTCAAAACCACA |
| AtActin | XM_021028231.1 | GGTAACATTGTGCTCAGTGGTGG | AACGACCTTAATCTTCATGCTGC |
| qPCR-AtDXS | NM_117647.3 | GAAGTCGCAAAGGGTATGACAAAGCA | CTGGATCAAATTTCACAACACCATGGTAT |
| qPCR-AtDXR | NM_125674.3 | TTCTGCCATATTTCAGTGTATTCAAGGTT | GCACAGATGAATCCTGTGTTTCAATC |
| qPCR-AtSQS1 | NM_119630.4 | GGAGAAGCAGATCCCTCCTG | AGTAGAACACACACGGCG |
| qPCR-AtSQS2 | NM_119631.3 | ATCTCAAAGCAAACCAATGTAAG | AGAACAACTGAAAAGGAAGAGAG |
| qPCR-AtCAS | NM_126681.3 | GACGGAGGTTGGGGTTTA | TGATTTAGTATCCAGTCTCGTCC |
| qPCR-AtIPI | NM_121649.6 | TGGGATCATGTTGAGAAAGGAAC | GTTGCCCAGTTTTGTCTGTAATCA |

图2 激素处理后大豆GmHMGR基因的表达模式分析 A:MeJA处理;B:ABA处理;数据为3个生物学重复的平均值 ± SE
Fig. 2 Expression patterns of GmHMGR gene in response to hormone treatment in soybean A: MeJA treatment; B: ABA treatment.The data represent the mean ± SE of three biological replicates
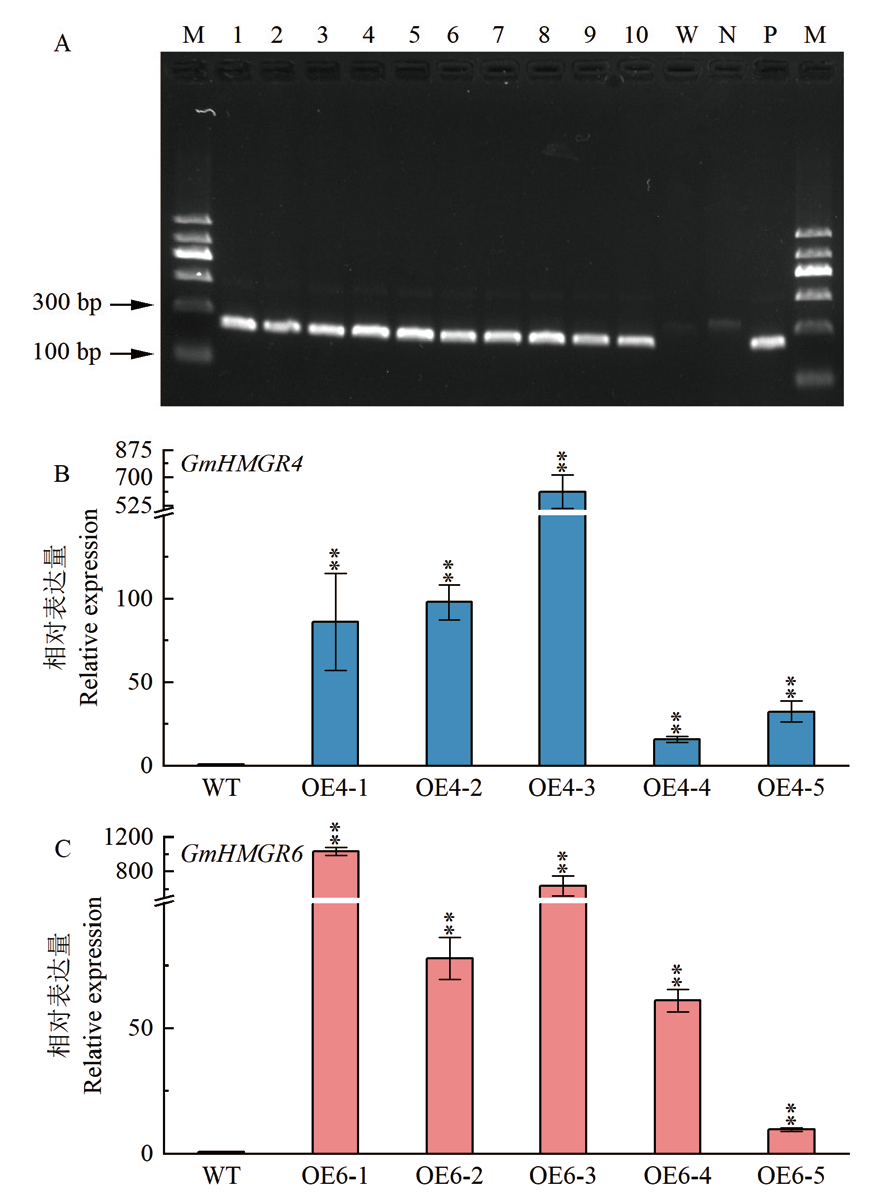
图4 T3代转基因拟南芥的鉴定 A:T3转基因拟南芥的PCR鉴定;M:DNA分子量标准;1-5:GmHMGR4转基因株系;6-10:GmHMGR6转基因株系;W:野生型对照;N:阴性对照;P:阳性对照;B:过量表达GmHMGR4转基因拟南芥的qPCR检测;C:过量表达GmHMGR6转基因拟南芥的qPCR检测。*表示0.05水平上显著差异,**表示0.01水平上显著差异。下同
Fig. 4 Identification of T3 transgenic A. thaliana A: Identification of T3 generation A. thaliana by PCR; M: DNA molecular weight marker. Lane 1-5: GmHMGR4 transgenic lines; lane 6-10: GmHMGR6 transgenic lines; W: wild-type control; N: negative control; P: positive control; B: qPCR detection of overexpressed GmHMGR4 transgenic A. thaliana; C: qPCR detection of overexpressed GmHMGR6 transgenic A. thaliana. * indicates significant difference at 0.05 level, and ** indicates significant difference at 0.01 level. The same below

图6 氧化和盐胁迫后转基因拟南芥的表型分析 A:野生型与转基因拟南芥在氧化和盐胁迫处理后的生长表型;B:拟南芥莲座直径统计;C、D:相对电导率与叶片损伤度的分析;E:MDA含量的测定
Fig. 6 Phenotypic analysis of transgenic A. thaliana after oxidative treatment and salt stress A: Growth phenotypes of wild-type and transgenic A. thaliana after oxidative and salt stress treatments. B: Size statistics of A. thaliana rosette diameter. C, D: Analysis of relative conductivity and leaf damage. E: Determination of MDA content
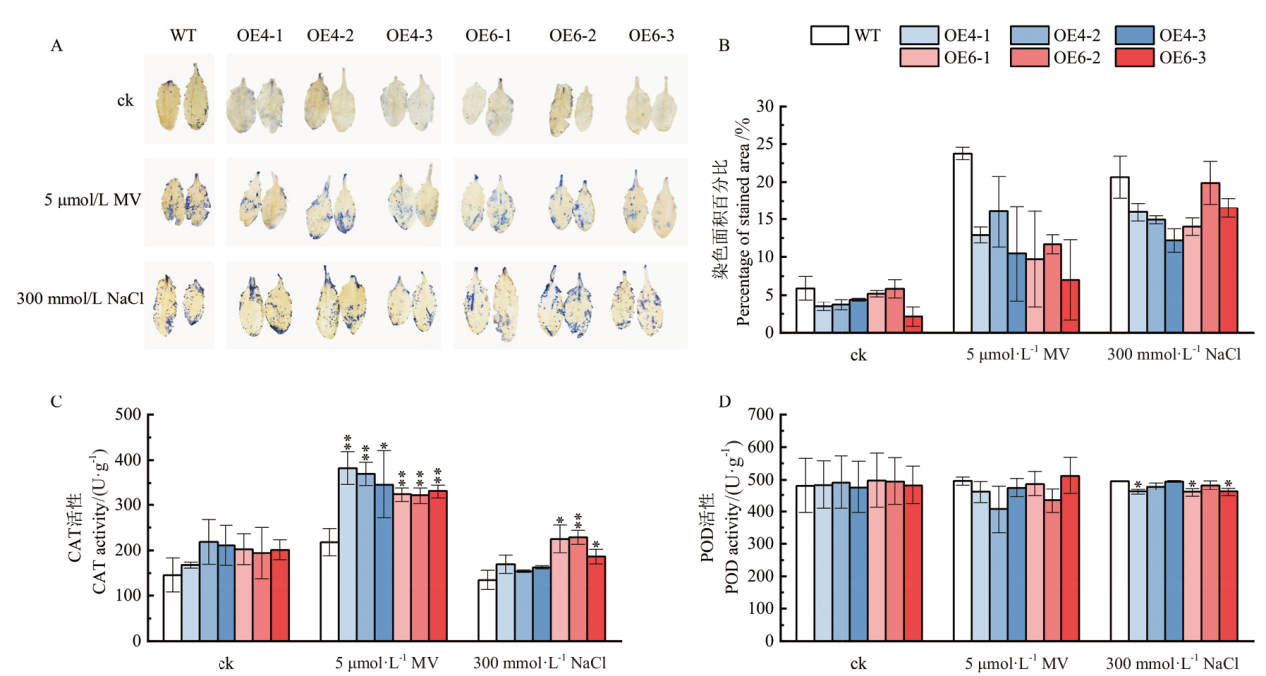
图7 氧化和盐胁迫处理后拟南芥的抗逆能力分析 A:拟南芥叶片在氧化和盐胁迫处理后的NBT染色;B:染色面积百分比的统计;C、D:CAT与POD活性的测定
Fig. 7 Stress-resistant ability analysis of transgenic A. thaliana after oxidative treatment and salt stress A: NBT staining of A. thaliana leaves after oxidative and salt stress treatments. B: Statistics of stained area percentage. C, D: Determination of CAT and POD activities
| [1] | 马靓, 丁鹏, 杨广笑, 等. 植物类萜生物合成途径及关键酶的研究进展[J]. 生物技术通报, 2006(S1): 22-30. |
| Ma L, Ding P, Yang GX, et al. Advances on the plant terpenoid isoprenoid biosynthetic pathway and its key enzymes[J]. Biotechnol Bull, 2006(S1): 22-30. | |
| [2] |
Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis[J]. Annu Rev Plant Biol, 2013, 64: 665-700.
doi: 10.1146/annurev-arplant-050312-120116 pmid: 23451776 |
| [3] |
Pulido P, Perello C, Rodriguez-Concepcion M. New insights into plant isoprenoid metabolism[J]. Mol Plant, 2012, 5(5): 964-967.
doi: 10.1093/mp/sss088 pmid: 22972017 |
| [4] |
Dong N, Ponciano G, McMahan CM, et al. Overexpression of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Parthenium argentatum(guayule)[J]. Ind Crops Prod, 2013, 46: 15-24.
doi: 10.1016/j.indcrop.2012.12.044 URL |
| [5] |
Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering[J]. Annu Rev Plant Biol, 2009, 60: 335-355.
doi: 10.1146/annurev.arplant.043008.091955 pmid: 19575586 |
| [6] |
Bick JA, Lange BM. Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane[J]. Arch Biochem Biophys, 2003, 415(2): 146-154.
doi: 10.1016/s0003-9861(03)00233-9 pmid: 12831836 |
| [7] |
陈妤, 朱沛煌, 李荣, 等. 植物异戊烯基转移酶研究进展[J]. 生物技术通报, 2021, 37(2): 149-161.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0476 URL |
|
Chen Y, Zhu PH, Li R, et al. Research progress of plant prenyltransferases[J]. Biotechnol Bull, 2021, 37(2): 149-161.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0476 URL |
|
| [8] |
Suzuki M, Kamide Y, Nagata N, et al. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1(HMG1)in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels[J]. Plant J, 2004, 37(5): 750-761.
doi: 10.1111/tpj.2004.37.issue-5 URL |
| [9] |
Rao S, Meng XX, Liao YL, et al. Characterization and functional analysis of two novel 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes(GbHMGR2 and GbHMGR3)from Ginkgo biloba[J]. Sci Rep, 2019, 9(1): 14109.
doi: 10.1038/s41598-019-50629-8 |
| [10] | 郝金凤, 李彤彤, 郭艳琼, 等. 甜瓜HMGR基因全长cDNA的克隆及序列分析[J]. 生物技术通报, 2010(8): 106-109, 115. |
| Hao JF, Li TT, Guo YQ, et al. Cloning and sequence analysis of the 3-hydroxy-3-methylglutaryl coenzyme-a reductase gene cDNA from melon cultivar Hetao(Cucumis melo L.cv.Hetao)[J]. Biotechnol Bull, 2010(8): 106-109, 115. | |
| [11] |
Liu W, Zhang ZQ, Li W, et al. Genome-wide identification and comparative analysis of the 3-hydroxy-3-methylglutaryl coenzyme a reductase(HMGR)gene family in Gossypium[J]. Molecules, 2018, 23(2): 193.
doi: 10.3390/molecules23020193 URL |
| [12] |
Kim YJ, Lee OR, Oh JY, et al. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng[J]. Plant Physiol, 2014, 165(1): 373-387.
doi: 10.1104/pp.113.222596 URL |
| [13] |
Zhang M, Liu H, Wang Q, et al. The 3-hydroxy-3-methylglutaryl-coenzyme A reductase 5 gene from Malus domestica enhances oxidative stress tolerance in Arabidopsis thaliana[J]. Plant Physiol Biochem, 2020, 146: 269-277.
doi: 10.1016/j.plaphy.2019.11.031 URL |
| [14] |
Choi D, Ward BL, Bostock RM. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid[J]. Plant Cell, 1992, 4(10): 1333-1344.
pmid: 1283354 |
| [15] |
Lumbreras V, Campos N, Boronat A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region[J]. Plant J, 1995, 8(4): 541-549.
doi: 10.1046/j.1365-313x.1995.8040541.x pmid: 7496400 |
| [16] |
Rupasinghe HPV, Almquist KC, Paliyath G, et al. Cloning of hmg1 and hmg2 cDNAs encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase and their expression and activity in relation to α-farnesene synthesis in apple[J]. Plant Physiol Biochem, 2001, 39(11): 933-947.
doi: 10.1016/S0981-9428(01)01316-X URL |
| [17] | 陈俞裴, 申甜, 邱飞, 等. 青蒿HMGR基因家族克隆及对茉莉酸甲酯和损伤的响应[J]. 西南大学学报: 自然科学版, 2017, 39(12): 44-51. |
| Chen YP, Shen T, Qiu F, et al. Molecular cloning and expression analysis of HMG-CoA(3-hydroxy-3-methylglutaryl-CoA)reductase gene family upon MeJA and mechanical injury treatments in Artemisia annua L[J]. J Southwest Univ Nat Sci, 2017, 39(12): 44-51. | |
| [18] | 刁秀楠, 斛如媛, 高春艳, 等. 大豆HMGR基因家族的鉴定与表达分析[J]. 山西农业大学学报: 自然科学版, 2019, 39(4)9-16 |
| Diao XN, Hu RY, Gao CY, et al. Identification and expression analysis of HMGR gene family in Glycine max[J]. J Shanxi Agric Univ Nat Sci Ed, 2019, 39(4)9-16 | |
| [19] | Abramoff M, Magalhães P, Ram SJ. Image processing with ImageJ[J]. Biophotonics Int, 2003, 11: 36-42. |
| [20] |
Bouvier F, Rahier A, Camara B. Biogenesis, molecular regulation and function of plant isoprenoids[J]. Prog Lipid Res, 2005, 44(6): 357-429.
doi: 10.1016/j.plipres.2005.09.003 pmid: 16289312 |
| [21] |
Jin HN, Song ZH, Nikolau BJ. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development[J]. Plant J, 2012, 70(6): 1015-1032.
doi: 10.1111/tpj.2012.70.issue-6 URL |
| [22] |
Liao P, Chen XJ, Wang MF, et al. Improved fruit α-tocopherol, carotenoid, squalene and phytosterol contents through manipulation of Brassica juncea 3-HYDROXY-3-METHYLGLUTARYL-COA SYNTHASE1 in transgenic tomato[J]. Plant Biotechnol J, 2018, 16(3): 784-796.
doi: 10.1111/pbi.12828 pmid: 28881416 |
| [23] | Liao P, Wang H, Wang MF, et al. Transgenic tobacco overexpressing Brassica juncea HMG-CoA synthase 1 shows increased plant growth, pod size and seed yield[J]. PLoS One, 2014, 9(5): e98264. |
| [24] |
Manzano D, Andrade P, Caudepón D, et al. Suppressing farnesyl diphosphate synthase alters chloroplast development and triggers sterol-dependent induction of jasmonate- and Fe-related responses[J]. Plant Physiol, 2016, 172(1): 93-117.
doi: 10.1104/pp.16.00431 pmid: 27382138 |
| [25] |
Kevei Z, Lougnon G, Mergaert P, et al. 3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula[J]. Plant Cell, 2007, 19(12): 3974-3989.
doi: 10.1105/tpc.107.053975 URL |
| [26] | 黄晶, 段续伟, 张文娜, 等. 杜梨HMGR基因克隆及其转基因烟草种子耐盐性分析[J]. 中国农业大学学报, 2015, 20(1): 60-67. |
| Huang J, Duan XW, Zhang WN, et al. Clone of pear HMGR gene and salt-tolerance analysis of its transgenic tobacco seed[J]. J China Agric Univ, 2015, 20(1): 60-67. | |
| [27] |
Lv DM, Zhang TT, Deng S, et al. Functional analysis of the Malus domestica MdHMGR2 gene promoter in transgenic Arabidopsis thaliana[J]. Biol Plant, 2016, 60(4): 667-676.
doi: 10.1007/s10535-016-0637-z URL |
| [28] |
Lv DM, Zhang YH. Isolation and functional analysis of apple MdHMGR1 and MdHMGR4 gene promoters in transgenic Arabi-dopsis thaliana[J]. Plant Cell Tiss Organ Cult, 2017, 129(1): 133-143.
doi: 10.1007/s11240-016-1162-7 URL |
| [29] |
Leivar P, Antolín-Llovera M, Ferrero S, et al. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A[J]. Plant Cell, 2011, 23(4): 1494-1511.
doi: 10.1105/tpc.110.074278 URL |
| [30] |
Lange I, Poirier BC, Herron BK, et al. Comprehensive assessment of transcriptional regulation facilitates metabolic engineering of isoprenoid accumulation in Arabidopsis[J]. Plant Physiol, 2015, 169(3): 1595-1606.
doi: 10.1104/pp.15.00573 pmid: 26282236 |
| [31] |
Nieto B, Forés O, Arró M, et al. Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways[J]. Phytochemistry, 2009, 70(1): 53-59.
doi: 10.1016/j.phytochem.2008.10.010 URL |
| [32] | Dai ZB, Cui GH, Zhou SF, et al. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation[J]. J Plant Physiol, 2011, 168(2): 148-157. |
| [33] |
Holmberg N, Harker M, Wallace AD, et al. Co-expression of N-terminal truncated 3-hydroxy-3-methylglutaryl CoA reductase and C24-sterol methyltransferase type 1 in transgenic tobacco enhances carbon flux towards end-product sterols[J]. Plant J, 2003, 36(1): 12-20.
pmid: 12974807 |
| [34] |
Laule O, Fürholz A, Chang HS, et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2003, 100(11): 6866-6871.
doi: 10.1073/pnas.1031755100 URL |
| [35] | Pallavi S, Bhushan JA, Shanker DR, et al. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions[J]. J Bot, 2012, 2012: 1-26. |
| [36] |
Wei H, Movahedi A, Xu C, et al. Overexpression of PtHMGR enhances drought and salt tolerance of poplar[J]. Ann Bot, 2020, 125(5): 785-803.
doi: 10.1093/aob/mcz158 URL |
| [37] |
Wang H, Nagegowda DA, Rawat R, et al. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance[J]. Plant Biotechnol J, 2012, 10(1): 31-42.
doi: 10.1111/j.1467-7652.2011.00631.x pmid: 21645203 |
| [38] |
Vickers CE, Gershenzon J, Lerdau MT, et al. A unified mechanism of action for volatile isoprenoids in plant abiotic stress[J]. Nat Chem Biol, 2009, 5(5): 283-291.
doi: 10.1038/nchembio.158 pmid: 19377454 |
| [39] |
Loreto F, Schnitzler JP. Abiotic stresses and induced BVOCs[J]. Trends Plant Sci, 2010, 15(3): 154-166.
doi: 10.1016/j.tplants.2009.12.006 pmid: 20133178 |
| [40] |
Loreto F, Dicke M, Schnitzler JP, et al. Plant volatiles and the environment[J]. Plant Cell Environ, 2014, 37(8): 1905-1908.
doi: 10.1111/pce.12369 URL |
| [1] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [2] | 魏茜雅, 秦中维, 梁腊梅, 林欣琪, 李映志. 褪黑素种子引发处理提高朝天椒耐盐性的作用机制[J]. 生物技术通报, 2023, 39(7): 160-172. |
| [3] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [4] | 马玉倩, 孙东辉, 岳浩峰, 辛佳瑜, 刘宁, 曹志艳. 具有辅助降解纤维素功能的大斑刚毛座腔菌糖苷水解酶GH61的鉴定、异源表达及功能分析[J]. 生物技术通报, 2023, 39(4): 124-135. |
| [5] | 陈楠楠, 王春来, 蒋振忠, 焦鹏, 关淑艳, 马义勇. 玉米ZmDHN15基因在烟草中的遗传转化及抗冷性分析[J]. 生物技术通报, 2023, 39(4): 259-267. |
| [6] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [7] | 王海龙, 李雨倩, 王勃, 邢国芳, 张杰伟. 谷子SiMAPK3基因的克隆和表达特性分析[J]. 生物技术通报, 2023, 39(3): 123-132. |
| [8] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [9] | 王琪, 胡哲, 富薇, 李光哲, 郝林. 伯克霍尔德氏菌GD17对黄瓜幼苗耐干旱的调节[J]. 生物技术通报, 2023, 39(3): 163-175. |
| [10] | 杜清洁, 周璐瑶, 杨思震, 张嘉欣, 陈春林, 李娟起, 李猛, 赵士文, 肖怀娟, 王吉庆. 过表达CaCP1提高转基因烟草对盐胁迫的敏感性[J]. 生物技术通报, 2023, 39(2): 172-182. |
| [11] | 陈奕博, 杨万明, 岳爱琴, 王利祥, 杜维俊, 王敏. 基于SLAF标记的大豆遗传图谱构建及苗期耐盐性QTL定位[J]. 生物技术通报, 2023, 39(2): 70-79. |
| [12] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [13] | 汪明滔, 刘建伟, 赵春钊. 植物调控盐胁迫下细胞壁完整性的分子机制[J]. 生物技术通报, 2023, 39(11): 18-27. |
| [14] | 周恒, 谢彦杰. 植物氧化胁迫信号应答的研究进展[J]. 生物技术通报, 2023, 39(11): 36-43. |
| [15] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||