








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 275-287.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0446
李明坤( ), 毕美营, 张天航, 吴翔宇, 杨培儒, 应明(
), 毕美营, 张天航, 吴翔宇, 杨培儒, 应明( )
)
收稿日期:2024-05-14
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
应明,女,博士,副教授,研究方向:基因工程与分子生物学;E-mail: ym@tjut.edu.cn作者简介:李明坤,男,研究方向:分子生物学;E-mail: MingkunLee@outlook.com
基金资助:
LI Ming-kun( ), BI Mei-ying, ZHANG Tian-hang, WU Xiang-yu, YANG Pei-ru, YING Ming(
), BI Mei-ying, ZHANG Tian-hang, WU Xiang-yu, YANG Pei-ru, YING Ming( )
)
Received:2024-05-14
Published:2024-10-26
Online:2024-11-20
摘要:
【目的】长期的化学耕作导致根系微生物的农用功能退化,以植物根部分离得到的一株无功能短小芽胞杆菌(Bacillus pumilus)为研究对象,探究恢复其农用功能的方法。【方法】采用一种新型的CRISPR/Cas多基因编辑系统UgRNA/Cas9,突变B. pumilus全基因组碳分解代谢物阻遏顺式作用元件CRE,改变该菌的碳源选择性。【结果】通过基因测序发现,LG3145的碳代谢和次级代谢途径的部分基因CRE位点发生了缺失、增添、转换、颠换等类型的突变。通过比较代谢组学和转录组学分析,LG3145所吸收的大部分碳源通过磷酸戊糖和氨基酸代谢途径进入了次级代谢通路,使细胞产生了细胞色素、表面活性素和杆菌素等非核糖体肽类抗菌素。经过UgRNA/Cas9编辑后,LG3145可以定殖于植物根系,促进小麦等农作物生长,提高植株的抗病菌能力。【结论】LG3145全基因组CRE序列的突变改变了菌体碳代谢流方向,使该细菌与植物之间形成了有益的互作关系。
李明坤, 毕美营, 张天航, 吴翔宇, 杨培儒, 应明. UgRNA/Cas9多基因编辑法恢复根际细菌农用功能的研究[J]. 生物技术通报, 2024, 40(10): 275-287.
LI Ming-kun, BI Mei-ying, ZHANG Tian-hang, WU Xiang-yu, YANG Pei-ru, YING Ming. Restoration of Agricultural Function of Rhizobacteria by UgRNA/Cas9 Multi-gene Editing[J]. Biotechnology Bulletin, 2024, 40(10): 275-287.
| 名称Name | UgRNA序列 UgRNA sequence(5'-3') | 相似度 Similarity/% |
|---|---|---|
| Cac: | AAACTGTAAGCGTTATCAATACGC | 70 |
| AAAAGCGTATTGATAACGCTTACA | ||
| Cpt : | AAACCGCAACCAAAGCTTCAGATT | 70 |
| AAAAAATCTGAAGCTTTGGTTGCG | ||
| Cax : | AAACGAAAGCGTTACCAGCAATAG | 55 |
| AAAACTATTGCTGGTAACGCTTTC |
表1 UgRNA寡核苷酸序列
Table 1 UgRNA oligonucleotide sequences
| 名称Name | UgRNA序列 UgRNA sequence(5'-3') | 相似度 Similarity/% |
|---|---|---|
| Cac: | AAACTGTAAGCGTTATCAATACGC | 70 |
| AAAAGCGTATTGATAACGCTTACA | ||
| Cpt : | AAACCGCAACCAAAGCTTCAGATT | 70 |
| AAAAAATCTGAAGCTTTGGTTGCG | ||
| Cax : | AAACGAAAGCGTTACCAGCAATAG | 55 |
| AAAACTATTGCTGGTAACGCTTTC |
| 质粒Plasmid | 特性Characterization | 来源Source |
|---|---|---|
| pCas9 | Bacterial expression of Cas9 nuclease, tracrRNA and crRNA guide, E.coli | Addgene( |
| pCas9-Cax | pCas9 derivate, containing crRNA of Cax | 本实验This study[ |
| pCas9-Cac | pCas9 derivate, containing crRNA of Cac | 本实验This study[ |
| pCas9-Cpt | pCas9 derivate, containing crRNA of Cpt | 本实验This study[ |
表2 实验所用质粒
Table 2 Plasmids used in this study
| 质粒Plasmid | 特性Characterization | 来源Source |
|---|---|---|
| pCas9 | Bacterial expression of Cas9 nuclease, tracrRNA and crRNA guide, E.coli | Addgene( |
| pCas9-Cax | pCas9 derivate, containing crRNA of Cax | 本实验This study[ |
| pCas9-Cac | pCas9 derivate, containing crRNA of Cac | 本实验This study[ |
| pCas9-Cpt | pCas9 derivate, containing crRNA of Cpt | 本实验This study[ |
| 引物Primer | 序列 Sequence(5'-3') | 引物Primer | 序列 Sequence(5'-3') | |
|---|---|---|---|---|
| glpFKD-F | GTGCTATGGAAAAAAGCATGGAGT | glpFKD-R | TAAATGCGCTCCGCTGATTCC | |
| ptsGHI-F | CTGCAGATTCTGGAATTCACGCAC | ptsGHI-R | AGATAGTGATTGTAGCACCTTTCGC | |
| ykgB-F | TCCACTGTTCCTTTTGGTCTTCC | ykgB-R | TTCTCAATCTGCTGCTTGTCCG | |
| crr-F | GCTCAAGCCCTGAGAGTGTT | crr-R | GCTCGAATACGAGGAAGTTGT | |
| fsa-F | CTGCACTGACAGAACCTGATACCA | fsa-R | GGAAGAAATCGGTAAAATGACAGGTC | |
| fbaA-F | ATGTAGCGTCCTGCTCCTTCA | fbaA-R | CCGTGTCATCATTATGACAGCC | |
| eno-F | CAGGATCGTTCTTTTCTTCACCA | eno-R | GAAGGCAGCATGGTGAAAGTC | |
| gap-F | GCATAACTGTGAGAAGAATCTGGC | gap-R | CTTGTTTTCCGTACACCGCTTT | |
| tkt-F | GAGACCGGTTTGTCAGACGC | tkt-R | GATGCTCAGGTTGTATCCGCTTA | |
| pdhABCD-F | CCAAGTTGAAGACAGTGAACCGT | pdhABCD-R | TACGACTTCGCCTTTTTCGTTT | |
| pgi-F | GGGCAAATTCTTCGCGATCA | pgi-R | CATACACTCGATCACAATGTGG | |
| guaB-F | TTCTCGATGTTACACCATGCGC | guaB-R | GATACCAAGACCACCTTGACGT | |
| tenA-F | CATGCTCAGGGAGTTTCCGTTTA | tenA-R | CAATATCTCTTCGAATCGTCCGC | |
| ribA-F | CCTGGTCGATGGATCAGGGA | ribA-R | GGTGCGAAGGCATACCACTTA | |
| ribBD-F | CGCTTAGGGCCTGTCAGAAG | ribBD-R | TTCCGCCACCTTTCAGTTCT | |
| crtNIM-F | ACGGAAGGAGCTTCCTTTCA | crtNIM-R | ATGAAAGCTGCGGAAGTGCT | |
| crtP-F | CAGGCTTCTATCCGTTTCTCCT | crtP-R | GGTATGGCTTTTTCCACAGGGG | |
| glk-F | CCATTCCAATCCCAATCAGCG | glk-R | GGAGAGACAGCGAAAGCAGAG | |
| glmS-F | GATCACCCGCTACATTGTGATTGAC | glmS-R | TTTGATGCGGATGCGCATTTAGA | |
| glmM-F | GAAGCAGCCGAATCTCGTCG | glmM-R | CGAGATACGCACTTCCCATC | |
| glmU-F | GGAGAGTTTCGCGACATTGCA | glmU-R | CCCAGCTGTTCTTCTTGAAGTG | |
| murA-F | GGCGAGCGGAGAAGATTCTT | murA-R | GAGGGAACCAGTTCTCCGTA | |
| murC-F | GGACAGTATACCCAGTATCATG | murC-R | CGAACAGGAGGAATTAATGCT | |
| murD-F | GGCTCAAATCGCTGTGAAATATG | murD-R | CCGTAACCAAGTGTGACAAATAAC | |
| murE-F | GGGATAGCCTGCCTGAGATG | murE-R | GCCTGTGATCCCTATGAGATTC | |
| murGB-F | GGAGAGGCATACGAATAGCACT | murGB-R | TAGAAGCCGCATAAACCACAGG | |
| accAD-F | GGTTTTCTGAGATCAGGTGC | accAD-R | AGTGTGCACGTTCTTTCTGA | |
| srfA-F | GCGGTATTTAGTCGATTATTTTCGG | srfA-R | AAGGGAGTCAGTTTGACGAATGTAC | |
| bacABCDF-F | GCAGGACTACTGATCATGGCT | bacABCDF-R | GCCGAGTGTAGTGGTGGAATG | |
| bacG-F | CTATTGCCGCAAAGGCCTCA | bacG-R | GCGTGAGGGTTCTCCTACAAAG | |
| aroA-F | CGCTGCTATGACACCTGACT | aroA-R | CCAACGTCTTTACGTATTACGGT | |
| aroBC-F | GCGCTGATTGCAGCACTTTATC | aroBC-R | GCCAAGGTGTTCATCAAATGAGC | |
| aroG-F | CTTCTTGAAGTTCTAGACCTGCTT | aroG-R | CAAGAGTTTGAAGCATTTGCGTCT | |
| liaGFRS-F | GCTTCCACCCGTAATCTTCACAT | liaGFRS-R | GCAGCAGTTAGAAACGAAGC | |
| liaHI-F | GCCACTTCACCATTGATCATCG | liaHI-R | GGCGCAGCTCTCTTACATGT |
表3 实验引物序列
Table 3 Primers used in this study
| 引物Primer | 序列 Sequence(5'-3') | 引物Primer | 序列 Sequence(5'-3') | |
|---|---|---|---|---|
| glpFKD-F | GTGCTATGGAAAAAAGCATGGAGT | glpFKD-R | TAAATGCGCTCCGCTGATTCC | |
| ptsGHI-F | CTGCAGATTCTGGAATTCACGCAC | ptsGHI-R | AGATAGTGATTGTAGCACCTTTCGC | |
| ykgB-F | TCCACTGTTCCTTTTGGTCTTCC | ykgB-R | TTCTCAATCTGCTGCTTGTCCG | |
| crr-F | GCTCAAGCCCTGAGAGTGTT | crr-R | GCTCGAATACGAGGAAGTTGT | |
| fsa-F | CTGCACTGACAGAACCTGATACCA | fsa-R | GGAAGAAATCGGTAAAATGACAGGTC | |
| fbaA-F | ATGTAGCGTCCTGCTCCTTCA | fbaA-R | CCGTGTCATCATTATGACAGCC | |
| eno-F | CAGGATCGTTCTTTTCTTCACCA | eno-R | GAAGGCAGCATGGTGAAAGTC | |
| gap-F | GCATAACTGTGAGAAGAATCTGGC | gap-R | CTTGTTTTCCGTACACCGCTTT | |
| tkt-F | GAGACCGGTTTGTCAGACGC | tkt-R | GATGCTCAGGTTGTATCCGCTTA | |
| pdhABCD-F | CCAAGTTGAAGACAGTGAACCGT | pdhABCD-R | TACGACTTCGCCTTTTTCGTTT | |
| pgi-F | GGGCAAATTCTTCGCGATCA | pgi-R | CATACACTCGATCACAATGTGG | |
| guaB-F | TTCTCGATGTTACACCATGCGC | guaB-R | GATACCAAGACCACCTTGACGT | |
| tenA-F | CATGCTCAGGGAGTTTCCGTTTA | tenA-R | CAATATCTCTTCGAATCGTCCGC | |
| ribA-F | CCTGGTCGATGGATCAGGGA | ribA-R | GGTGCGAAGGCATACCACTTA | |
| ribBD-F | CGCTTAGGGCCTGTCAGAAG | ribBD-R | TTCCGCCACCTTTCAGTTCT | |
| crtNIM-F | ACGGAAGGAGCTTCCTTTCA | crtNIM-R | ATGAAAGCTGCGGAAGTGCT | |
| crtP-F | CAGGCTTCTATCCGTTTCTCCT | crtP-R | GGTATGGCTTTTTCCACAGGGG | |
| glk-F | CCATTCCAATCCCAATCAGCG | glk-R | GGAGAGACAGCGAAAGCAGAG | |
| glmS-F | GATCACCCGCTACATTGTGATTGAC | glmS-R | TTTGATGCGGATGCGCATTTAGA | |
| glmM-F | GAAGCAGCCGAATCTCGTCG | glmM-R | CGAGATACGCACTTCCCATC | |
| glmU-F | GGAGAGTTTCGCGACATTGCA | glmU-R | CCCAGCTGTTCTTCTTGAAGTG | |
| murA-F | GGCGAGCGGAGAAGATTCTT | murA-R | GAGGGAACCAGTTCTCCGTA | |
| murC-F | GGACAGTATACCCAGTATCATG | murC-R | CGAACAGGAGGAATTAATGCT | |
| murD-F | GGCTCAAATCGCTGTGAAATATG | murD-R | CCGTAACCAAGTGTGACAAATAAC | |
| murE-F | GGGATAGCCTGCCTGAGATG | murE-R | GCCTGTGATCCCTATGAGATTC | |
| murGB-F | GGAGAGGCATACGAATAGCACT | murGB-R | TAGAAGCCGCATAAACCACAGG | |
| accAD-F | GGTTTTCTGAGATCAGGTGC | accAD-R | AGTGTGCACGTTCTTTCTGA | |
| srfA-F | GCGGTATTTAGTCGATTATTTTCGG | srfA-R | AAGGGAGTCAGTTTGACGAATGTAC | |
| bacABCDF-F | GCAGGACTACTGATCATGGCT | bacABCDF-R | GCCGAGTGTAGTGGTGGAATG | |
| bacG-F | CTATTGCCGCAAAGGCCTCA | bacG-R | GCGTGAGGGTTCTCCTACAAAG | |
| aroA-F | CGCTGCTATGACACCTGACT | aroA-R | CCAACGTCTTTACGTATTACGGT | |
| aroBC-F | GCGCTGATTGCAGCACTTTATC | aroBC-R | GCCAAGGTGTTCATCAAATGAGC | |
| aroG-F | CTTCTTGAAGTTCTAGACCTGCTT | aroG-R | CAAGAGTTTGAAGCATTTGCGTCT | |
| liaGFRS-F | GCTTCCACCCGTAATCTTCACAT | liaGFRS-R | GCAGCAGTTAGAAACGAAGC | |
| liaHI-F | GCCACTTCACCATTGATCATCG | liaHI-R | GGCGCAGCTCTCTTACATGT |
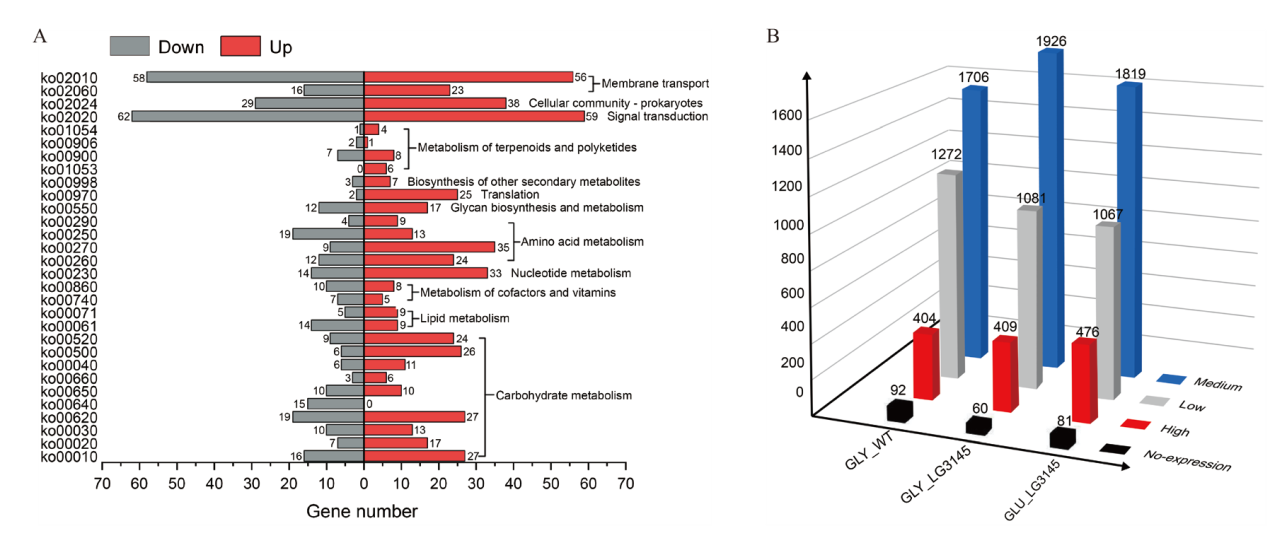
图1 B. pumilus LG3145的比较转录组学分析 A:GYMM中LG3145转录组KEGG代谢通路富集分析。Down:下调;Up:上调。B:菌株LG3145全基因组表达量对比分析。GLY_WT:GUMM中野生菌WT;GLY_LG3145:GYMM中LG3145;GLU_LG3145:GUMM中LG3145。High:高表达(FPKM ≥ 900);Medium:中等表达(500 ≤ FPKM < 900);Low:低表达(50 ≤ FPKM < 500),No-expression:无表达(1 < FPKM < 50)
Fig. 1 Comparative transcriptomic analysis of B. pumilus LG3145 A: Enriching analysis in KEGG metabolic pathways of strain LG3145 in GYMM. Down: Downregulated; Up: upregulated. B: Eomparative analysis of whole-genome expression of strain LG3145. GLY_WT: WT in GUMM; GLY_LG3145: LG3145 in GYMM; GLU_LG3145: LG3145 in GUMM. High: High expression(FPKM ≥ 900). Medium: Medium expression(500 ≤ FPKM < 900). Low: Low expression(50 ≤ FPKM < 500). No-expression: No expression(1 < FPKM < 50)
| Gene or operon | cre-like and flanking sequenceab | TxSSc | Gene or operon | cre-like and flanking sequenceab | TxSSc | |
|---|---|---|---|---|---|---|
| ackA | TATCG ATGTAAGCGGTAACA[0,0]GTTCA | -52 | sucC | TTAAA ATGAAAGCGCAGTCT[0,0]ATTTT | -6 | |
| glpFKD | AGAGG ATGAAAACGTTGTCA[0,0]ATAAA | -190 | glk | CAATT TTAAAGCGAAACA[2,0]ATATA | -330 | |
| ATTTA AAGAAATCCTCGCA[1,0]TATCA | 81 | glmS | TGTTG ATGCAAACGTAGCT[1,0]CTTCA | -259 | ||
| ptsGHI | TAACT ATGAAATGATGTCA[1,0]GGGAG | -66 | glmM | CTCTT TAGGACGCTCTT[3,0]TTCTA | -191 | |
| ykgB | AAATA TCAAACGGTTTCT[0,2]CAAAT | -148 | glmU | ACAGC ATTTAAAGAAACA[0,2]TCCAT | -157 | |
| crr | CGTGT ATGAAACTACGCT[0,2]CATAC | -45 | murA | CAAGG AAGAAATGCTGACT[0,1]GCAGA | -189 | |
| fsa | ATCTT AGAAAGCGCAATCA[1,0]AATTT | -59 | TGCAA TAGCAATGGTAATGA[0,2]GCATC | -214 | ||
| fbaA | AGGTT TTGCAAAGTGTCA[0,2]AAGCA | -261 | murC | CAATT TTGGAATGGACGCT[1,0]TTTCG | -248 | |
| ATCCA AAGAATCGGTGCT[2,0]TGACA | -276 | mraY/murD | CCATC AAAAATGCATTCA[0,2]TTATT | -56 | ||
| eno/gpm/tpi/pgk | AAATA AAAAACCGTTCACA[1,0]GTTTT | 211 | murE | AGCCA TTGTACGAACAC[3,0]CTCTT | 193 | |
| gap/cggR | AAGAG TTGAAAAGAAAGT[2,0]TATCC | -130 | murGB | TTGTT TAGAAAACGTTAT[2,0]TTGAC | -110 | |
| CATGG ATGAATCTTCAT[3,0]ATAGA | -255 | TTATT TTGCAAAGTATAA[2,0]AAAAT | -161 | |||
| tkt | GATAA TAAAAACGGTCTCT[0,1]GTTAA | 171 | TGCTG AAGCAAGAAACCA[2,0]TGTTG | -190 | ||
| pdhABCD | ATGTT TGAAACGATCCCA[2,0]GTGGC | -127 | pyk/pfkA/accAD | GACAT TTGCAAAGAATGCA[1,0]AAACA | 189 | |
| pgi | GGAAC TAGCATGAAATCA[0,2]AGTAA | -67 | srfA | GGAGG ATGAAAAGAATACA[1,0]GTTTA | -6 | |
| GGCTT TTAAATCGCTAAA[0,2]TGTCA | -155 | bacABCDF | ACACA TTGAAACGTAAGCT[1,0]CCATT | -161 | ||
| guaB | AAATA AAAAACCGTTCACA[1,0]GTTTT | -259 | bacG | AAACA AAGGAACCGGTGTCT[0,0]CTTTT | 189 | |
| tenA | GTCGT ATGTAAGCTCACT[0,2]AATCG | 183 | AGGGA TTGGAAAGCAATC[2,0]CCATA | 70 | ||
| ribA | CGACT AGGAAGCGCAGTA[0,2]TGGTG | -83 | GTGTA CGATGGCCATGAT[0,2]CCGCT | -235 | ||
| ribBD | ATAAA AAGGCAAGACGCT[0,2]CAAAG | 58 | aroA | CATTG TACCAACTCATCA[2,0]TATAA | -68 | |
| crtNIM | AACGA TAGAAACGCAAAT[2,0]GGATC | 389 | aroBC | GAGAT TGAAGCGTTCCG[2,0]GCTGC | 126 | |
| TCGTC TTGGAAACAATCT[0,2]ATACA | 273 | GCGCA ATGAAACGAAGTT[2,0]AACGT | 77 | |||
| AATGT AAGAAAGCGCCCT[2,0]TTATT | -145 | aroG | GATCA ATGAAAGGGAAACA[1,0]TTGTA | 73 | ||
| crtP | AAGAA TTTAAACGATCCA[2,0]AGGAC | -71 | liaGFRS | AAACT AAGAAATCGAGACA[0,1]GGTCG | -113 | |
| AAGGA TATGACGCGATCT[2,0]TGGCG | -165 | liaHI | ATCCA ATAAAAGCACTCT[0,2]TCAAT | -143 | ||
| Consensus WWGNAANCGNWNNCW(W:A /T, N:A/T/G/C) | ||||||
表4 对B. pumilus WT中的部分操纵子的CRE位点的预测
Table 4 Prediction of CRE sites in some operons in B. pumilus WT
| Gene or operon | cre-like and flanking sequenceab | TxSSc | Gene or operon | cre-like and flanking sequenceab | TxSSc | |
|---|---|---|---|---|---|---|
| ackA | TATCG ATGTAAGCGGTAACA[0,0]GTTCA | -52 | sucC | TTAAA ATGAAAGCGCAGTCT[0,0]ATTTT | -6 | |
| glpFKD | AGAGG ATGAAAACGTTGTCA[0,0]ATAAA | -190 | glk | CAATT TTAAAGCGAAACA[2,0]ATATA | -330 | |
| ATTTA AAGAAATCCTCGCA[1,0]TATCA | 81 | glmS | TGTTG ATGCAAACGTAGCT[1,0]CTTCA | -259 | ||
| ptsGHI | TAACT ATGAAATGATGTCA[1,0]GGGAG | -66 | glmM | CTCTT TAGGACGCTCTT[3,0]TTCTA | -191 | |
| ykgB | AAATA TCAAACGGTTTCT[0,2]CAAAT | -148 | glmU | ACAGC ATTTAAAGAAACA[0,2]TCCAT | -157 | |
| crr | CGTGT ATGAAACTACGCT[0,2]CATAC | -45 | murA | CAAGG AAGAAATGCTGACT[0,1]GCAGA | -189 | |
| fsa | ATCTT AGAAAGCGCAATCA[1,0]AATTT | -59 | TGCAA TAGCAATGGTAATGA[0,2]GCATC | -214 | ||
| fbaA | AGGTT TTGCAAAGTGTCA[0,2]AAGCA | -261 | murC | CAATT TTGGAATGGACGCT[1,0]TTTCG | -248 | |
| ATCCA AAGAATCGGTGCT[2,0]TGACA | -276 | mraY/murD | CCATC AAAAATGCATTCA[0,2]TTATT | -56 | ||
| eno/gpm/tpi/pgk | AAATA AAAAACCGTTCACA[1,0]GTTTT | 211 | murE | AGCCA TTGTACGAACAC[3,0]CTCTT | 193 | |
| gap/cggR | AAGAG TTGAAAAGAAAGT[2,0]TATCC | -130 | murGB | TTGTT TAGAAAACGTTAT[2,0]TTGAC | -110 | |
| CATGG ATGAATCTTCAT[3,0]ATAGA | -255 | TTATT TTGCAAAGTATAA[2,0]AAAAT | -161 | |||
| tkt | GATAA TAAAAACGGTCTCT[0,1]GTTAA | 171 | TGCTG AAGCAAGAAACCA[2,0]TGTTG | -190 | ||
| pdhABCD | ATGTT TGAAACGATCCCA[2,0]GTGGC | -127 | pyk/pfkA/accAD | GACAT TTGCAAAGAATGCA[1,0]AAACA | 189 | |
| pgi | GGAAC TAGCATGAAATCA[0,2]AGTAA | -67 | srfA | GGAGG ATGAAAAGAATACA[1,0]GTTTA | -6 | |
| GGCTT TTAAATCGCTAAA[0,2]TGTCA | -155 | bacABCDF | ACACA TTGAAACGTAAGCT[1,0]CCATT | -161 | ||
| guaB | AAATA AAAAACCGTTCACA[1,0]GTTTT | -259 | bacG | AAACA AAGGAACCGGTGTCT[0,0]CTTTT | 189 | |
| tenA | GTCGT ATGTAAGCTCACT[0,2]AATCG | 183 | AGGGA TTGGAAAGCAATC[2,0]CCATA | 70 | ||
| ribA | CGACT AGGAAGCGCAGTA[0,2]TGGTG | -83 | GTGTA CGATGGCCATGAT[0,2]CCGCT | -235 | ||
| ribBD | ATAAA AAGGCAAGACGCT[0,2]CAAAG | 58 | aroA | CATTG TACCAACTCATCA[2,0]TATAA | -68 | |
| crtNIM | AACGA TAGAAACGCAAAT[2,0]GGATC | 389 | aroBC | GAGAT TGAAGCGTTCCG[2,0]GCTGC | 126 | |
| TCGTC TTGGAAACAATCT[0,2]ATACA | 273 | GCGCA ATGAAACGAAGTT[2,0]AACGT | 77 | |||
| AATGT AAGAAAGCGCCCT[2,0]TTATT | -145 | aroG | GATCA ATGAAAGGGAAACA[1,0]TTGTA | 73 | ||
| crtP | AAGAA TTTAAACGATCCA[2,0]AGGAC | -71 | liaGFRS | AAACT AAGAAATCGAGACA[0,1]GGTCG | -113 | |
| AAGGA TATGACGCGATCT[2,0]TGGCG | -165 | liaHI | ATCCA ATAAAAGCACTCT[0,2]TCAAT | -143 | ||
| Consensus WWGNAANCGNWNNCW(W:A /T, N:A/T/G/C) | ||||||

图2 以甘油为碳源时LG3145差异代谢物和差异基因KEGG代谢通路富集分析 A:差异代谢物代谢通路富集分析;B:差异基因代谢通路富集分析。气泡大小反应显著程度,红色反向箭头表示相同的富集途径。同一方向的蓝色和绿色箭头表示相关的代谢途径(P-value < 0.05,|log2 fold-change(FC)| ≥ 1)
Fig. 2 KEGG enrichment pathways analysis of altered metabolites and genes when LG3145 used glycerol as carbon source A: Enriched altered metabolite pathways. B: Enriched altered gene pathways. The bubble size indicates significance. The red reverse arrow indicates the same enrichment pathway. The blue and green arrows in the same direction indicate associated metabolic pathways.(P-value < 0.05,|log2 fold-change(FC)| ≥ 1)

图3 CRE位点测序分析图 :CRE位点; :与共识序列“ WWGNAANCGNWNNCW (W:A/T, N:A/T/G/C) ”不同的突变碱基或错配位点; :转录方向
Fig. 3 Chromatograms of the DNA sequencing of CRE sites : CRE sites; : distinct mutation bases or mismatch site with the consenus sequences “ WWGNAANCGNWNNCW (W:A/T, N:A/T/G/C) ”; : the direction of transcription
| 病原体Pathogen | 直径Diameter/cm | 抑制率 Inhibition/% | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | |||||||||||||
| EG | CK1 | CK2 | EG | CK1 | CK2 | EG | CK1 | CK2 | *CK | **EG | |||||
| F. Sheld | 0.70±0.07 | 0.70±0.02 | 0.70±0.03 | 0.80±0.06 | 1.25±0.05 | 0.96±0.05 | 0.95±0.04 | 2.10±0.13 | 1.15±0.08 | 45 | 55 | ||||
| H. Pass | 0.70±0.06 | 0.75±0.14 | 0.75±0.05 | 0.80±0.04 | 2.10±0.04 | 2.03±0.09 | 0.80±0.11 | 2.75±0.05 | 2.45±0.07 | 11 | 71 | ||||
| S. Weber | 0.60±0.13 | 0.75±0.10 | 0.68±0.11 | 0.65±0.12 | 1.55±0.08 | 1.65±0.06 | 0.75±0.12 | 3.30±0.07 | 2.90±0.04 | 12 | 77 | ||||
| B. Pers | 0.75±0.11 | 3.25±0.11 | 1.19±0.16 | 1.15±0.09 | 4.80±0.12 | 3.43±0.03 | 1.45±0.07 | 6.90±0.09 | 4.21±0.09 | 39 | 79 | ||||
| F. graminearum | 1.10±0.10 | 1.10±0.13 | 1.10±0.15 | 1.95±0.13 | 3.35±0.05 | 1.69±0.12 | 2.50±0.10 | 5.80±0.03 | 3.77±0.05 | 35 | 57 | ||||
| R. solaniKühn | 0.85±0.12 | 1.70±0.12 | 0.95±0.04 | 0.90±0.15 | 2.50±0.07 | 1.24±0.07 | 1.25±0.06 | 3.10±0.10 | 2.36±0.03 | 24 | 60 | ||||
| Average inhibition | 28 | 67 | |||||||||||||
表5 LG3145 对真菌病原体的抑制作用
Table 5 Inhibition to fungal pathogens by LG3145
| 病原体Pathogen | 直径Diameter/cm | 抑制率 Inhibition/% | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | |||||||||||||
| EG | CK1 | CK2 | EG | CK1 | CK2 | EG | CK1 | CK2 | *CK | **EG | |||||
| F. Sheld | 0.70±0.07 | 0.70±0.02 | 0.70±0.03 | 0.80±0.06 | 1.25±0.05 | 0.96±0.05 | 0.95±0.04 | 2.10±0.13 | 1.15±0.08 | 45 | 55 | ||||
| H. Pass | 0.70±0.06 | 0.75±0.14 | 0.75±0.05 | 0.80±0.04 | 2.10±0.04 | 2.03±0.09 | 0.80±0.11 | 2.75±0.05 | 2.45±0.07 | 11 | 71 | ||||
| S. Weber | 0.60±0.13 | 0.75±0.10 | 0.68±0.11 | 0.65±0.12 | 1.55±0.08 | 1.65±0.06 | 0.75±0.12 | 3.30±0.07 | 2.90±0.04 | 12 | 77 | ||||
| B. Pers | 0.75±0.11 | 3.25±0.11 | 1.19±0.16 | 1.15±0.09 | 4.80±0.12 | 3.43±0.03 | 1.45±0.07 | 6.90±0.09 | 4.21±0.09 | 39 | 79 | ||||
| F. graminearum | 1.10±0.10 | 1.10±0.13 | 1.10±0.15 | 1.95±0.13 | 3.35±0.05 | 1.69±0.12 | 2.50±0.10 | 5.80±0.03 | 3.77±0.05 | 35 | 57 | ||||
| R. solaniKühn | 0.85±0.12 | 1.70±0.12 | 0.95±0.04 | 0.90±0.15 | 2.50±0.07 | 1.24±0.07 | 1.25±0.06 | 3.10±0.10 | 2.36±0.03 | 24 | 60 | ||||
| Average inhibition | 28 | 67 | |||||||||||||

图4 菌株LG3145对植物病原菌抑制实验 A:LG3145实验组;B:空白对照组;C:野生菌WT对照组。1:串珠镰刀菌;2:长蠕孢菌;3:茄匐柄霉;4:灰葡萄孢菌;5:禾谷镰孢菌;6:立枯丝核菌
Fig. 4 Inhibition assay of strain LG3145 to plant pathogens A: LG3145 experimental group; B: Blank control group; C: Wild-type strain control group. 1: Fusarium moniliforme Sheldon; 2: Helminthosporium turcicum Pass;3: Stemphylium solani Weber; 4: Botrytis cinerea Pers; 5: Fusarium graminearum;6: Rhizoctonia solani Kühn

图5 菌株LG3145对小麦植株生长发育的影响 A:菌株LG3145和WT的冻末粉;B:小麦栽培对比实验;C:9 d后小麦苗对比分析。CK1:使用无菌水浇灌的空白组;CK2:使用WT菌剂浇灌的对照组
Fig. 5 Effects of strain LG3145 on the growth and development of wheat plants A: Freezedried powder of strain LG3145 and WT. B: Wheat cultivation comparative experiment. C: Comparative analysis of wheat shoots after 9 days. CK1: Blank control group with sterile water-irrigation; CK2: WT-irrigated control group
| 方法Treatment | 株高Plant height/cm | 株重Shoot weight/g | |||||
|---|---|---|---|---|---|---|---|
| Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | ||
| LG3145 | 2.21±0.11 | 5.83±0.17** | 7.63±0.22** | 9.81±0.52** | 11.81±0.44** | 12.61±0.46** | 2.81±0.11** |
| CK1 | 2.21±0.11 | 4.12±0.30 | 5.23±0.33 | 7.04±0.28 | 8.82±0.19 | 9.62±0.22 | 2.33±0.07 |
| CK2 | 2.13±0.09 | 4.81±0.33* | 6.21±0.44* | 8.42±0.54* | 10.01±0.48* | 11.03±0.49** | 2.65±0.09** |
表6 LG3145 对小麦幼苗生物学特性的影响
Table 6 Effects of LG3145 on biological characteristics of wheat seedlings
| 方法Treatment | 株高Plant height/cm | 株重Shoot weight/g | |||||
|---|---|---|---|---|---|---|---|
| Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | ||
| LG3145 | 2.21±0.11 | 5.83±0.17** | 7.63±0.22** | 9.81±0.52** | 11.81±0.44** | 12.61±0.46** | 2.81±0.11** |
| CK1 | 2.21±0.11 | 4.12±0.30 | 5.23±0.33 | 7.04±0.28 | 8.82±0.19 | 9.62±0.22 | 2.33±0.07 |
| CK2 | 2.13±0.09 | 4.81±0.33* | 6.21±0.44* | 8.42±0.54* | 10.01±0.48* | 11.03±0.49** | 2.65±0.09** |
| [1] | Schippers B, Bakker AW, Bakker PM. Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices[J]. Annu Rev Phytopathol, 1987, 25: 339-358. |
| [2] | Zhang N, Nunan N, Hirsch PR, et al. Theory of microbial coexistence in promoting soil-plant ecosystem health[J]. Biol Fertil Soils, 2021, 57(7): 897-911. |
| [3] |
Gao CX. Genome engineering for crop improvement and future agriculture[J]. Cell, 2021, 184(6): 1621-1635.
doi: 10.1016/j.cell.2021.01.005 pmid: 33581057 |
| [4] | Zhang XJ, Huang YJ, Harvey PR, et al. Enhancing plant disease suppression by Burkholderia vietnamiensis through chromosomal integration of Bacillus subtilis chitinase gene Chi113[J]. Biotechnol Lett, 2012, 34(2): 287-293. |
| [5] | Peng G, Zhao X, Li Y, et al. Engineering Bacillus velezensis with high production of acetoin primes strong induced systemic resistance in Arabidopsis thaliana[J]. Microbiol Res, 2019, 227: 126297. |
| [6] | Yi YL, Li ZB, Song CX, et al. Exploring plant-microbe interactions of the rhizobacteria Bacillus subtilis and Bacillus mycoides by use of the CRISPR-Cas9 system[J]. Environ Microbiol, 2018, 20(12): 4245-4260. |
| [7] |
Liu X, Wu SR, Xu J, et al. Application of CRISPR/Cas9 in plant biology[J]. Acta Pharm Sin B, 2017, 7(3): 292-302.
doi: 10.1016/j.apsb.2017.01.002 pmid: 28589077 |
| [8] | 周桓, 邵艳娜, 王涓, 等. 基于CRISPR/Cas技术的核酸检测研究进展[J]. 微生物学报, 2021, 61(12): 3856-3869. |
| Zhou H, Shao YN, Wang J, et al. Research progress of nucleic acid detection based on CRISPR/Cas technology[J]. Acta Microbiol Sin, 2021, 61(12): 3856-3869. | |
| [9] |
Walker TS, Bais HP, Grotewold E, et al. Root exudation and rhizosphere biology[J]. Plant Physiol, 2003, 132(1): 44-51.
doi: 10.1104/pp.102.019661 pmid: 12746510 |
| [10] |
Szilagyi-Zecchin VJ, Ikeda AC, Hungria M, et al. Identification and characterization of endophytic bacteria from corn(Zea mays L.) roots with biotechnological potential in agriculture[J]. AMB Express, 2014, 4: 26.
doi: 10.1186/s13568-014-0026-y pmid: 24949261 |
| [11] | Franzino T, Boubakri H, Cernava T, et al. Implications of carbon catabolite repression for plant-microbe interactions[J]. Plant Commun, 2021, 3(2): 100272. |
| [12] |
Nicholson WL, Park YK, Henkin TM, et al. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence[J]. J Mol Biol, 1987, 198(4): 609-618.
pmid: 3123701 |
| [13] | Wang YX, Cao LF, Bi MY, et al. Wobble editing of Cre-box by unspecific CRISPR/Cas9 causes CCR release and phenotypic changes in Bacillus pumilus[J]. Front Chem, 2021, 9: 717609. |
| [14] | 陈启民, 耿运琪, 倪津, 等. 短小芽胞杆菌作为芽胞杆菌属基因工程受体菌的研究[J]. 遗传学报, 1989, 16(3): 206-212. |
| Chen QM, Geng YQ, Ni J, et al. Bacillus subtilis as a receptor bacterium for genetic engineering of the genus Bacillus[J]. Journal of Genetics, 1989, 16(3): 206-212. | |
| [15] |
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2[J]. Nat Methods, 2012, 9(4): 357-359.
doi: 10.1038/nmeth.1923 pmid: 22388286 |
| [16] |
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12: 323.
doi: 10.1186/1471-2105-12-323 pmid: 21816040 |
| [17] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| [18] |
Wang LK, Feng ZX, Wang X, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data[J]. Bioinformatics, 2010, 26(1): 136-138.
doi: 10.1093/bioinformatics/btp612 pmid: 19855105 |
| [19] |
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data[J]. Bioinformatics, 2010, 26(1): 139-140.
doi: 10.1093/bioinformatics/btp616 pmid: 19910308 |
| [20] |
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Res, 2000, 28(1): 27-30.
doi: 10.1093/nar/28.1.27 pmid: 10592173 |
| [21] |
Miwa Y, Nakata A, Ogiwara A, et al. Evaluation and characterization of catabolite-responsive elements(cre)of Bacillus subtilis[J]. Nucleic Acids Res, 2000, 28(5): 1206-1210.
pmid: 10666464 |
| [22] | Rojas-Pirela M, Andrade-Alviárez D, Rojas V, et al. Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea[J]. Open Biol, 2020, 10(11): 200302. |
| [23] | Wu Q, Shabbir MAB, Peng DP, et al. Microbiological inhibition-based method for screening and identifying of antibiotic residues in milk, chicken egg and honey[J]. Food Chem, 2021, 363: 130074. |
| [24] |
Kotta-Loizou I. Mycoviruses and their role in fungal pathogenesis[J]. Curr Opin Microbiol, 2021, 63: 10-18.
doi: 10.1016/j.mib.2021.05.007 pmid: 34102567 |
| [25] | Ahsan T, Tian PC, Gao J, et al. Effects of microbial agent and microbial fertilizer input on soil microbial community structure and diversity in a peanut continuous cropping system[J]. J Adv Res, 2023: S 2090-S1232(23)00367-3. |
| [26] | Zhang JL, Li CX, Zhang WJ, et al. Wheat plant height locus RHT25 encodes a PLATZ transcription factor that interacts with DELLA(RHT1)[J]. Proc Natl Acad Sci USA, 2023, 120(19): e2300203120. |
| [27] |
李元召, 孙俊良, 梁新红, 等. 重组枯草芽胞杆菌PaE菌株的构建及碳代谢阻遏效应研究[J]. 食品科学, 2014, 35(23): 134-138.
doi: 10.7506/spkx1002-6630-201423027 |
| Li YZ, Sun JL, Liang XH, et al. Construction and carbon catabolite repression of recombinant Bacillus subtilis PaE[J]. Food Sci, 2014, 35(23): 134-138. | |
| [28] | Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization[J]. Soil Biol Biochem, 2010, 42(5): 669-678. |
| [29] | 邓嘉雯, 高云雨, 刘旭坤, 等. 多靶向沉默里氏木霉碳代谢阻遏物对纤维素酶活性和表达的调控研究[J]. 微生物学报, 2019, 59(4): 730-743. |
| Deng JW, Gao YY, Liu XK, et al. Cellulase activities and expression regulation by multiple targeted silencing carbon catabolic repressors in Trichoderma reesei[J]. Acta Microbiol Sin, 2019, 59(4): 730-743. | |
| [30] |
Chen C, Lu YQ, Wang LL, et al. CcpA-dependent carbon catabolite repression regulates fructooligosaccharides metabolism in Lactobacillus plantarum[J]. Front Microbiol, 2018, 9: 1114.
doi: 10.3389/fmicb.2018.01114 pmid: 29896178 |
| [31] | Huang YF, Lemieux MJ, Song JM, et al. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli[J]. Science, 2003, 301(5633): 616-620. |
| [32] |
Kimata K, Tanaka Y, Inada T, et al. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli[J]. EMBO J, 2001, 20(13): 3587-3595.
pmid: 11432845 |
| [33] | 董悦, 胡坤乐, 李兴林, 等. 代谢工程改造甘油代谢途径提高β-胡萝卜素产量[J]. 生物工程学报, 2017, 33(2): 247-260. |
|
Dong Y, Hu KL, Li XL, et al. Improving β-carotene production in Escherichia coli by metabolic engineering of glycerol utilization pathway[J]. Chinese Journal of Biotechnology, 2017, 33(2): 247-260.
doi: 10.13345/j.cjb.160275 pmid: 28956381 |
| [1] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [2] | 岳丽昕, 王清华, 刘泽洲, 孔素萍, 高莉敏. 基于转录组和WGCNA筛选大葱雄性不育相关基因[J]. 生物技术通报, 2024, 40(9): 212-224. |
| [3] | 聂祝欣, 郭瑾, 乔子洋, 李微薇, 张学燕, 刘春阳, 王静. 黑果枸杞不同发育时期果实花色苷合成的转录组分析[J]. 生物技术通报, 2024, 40(8): 106-117. |
| [4] | 周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151. |
| [5] | 王睿, 戚继. 整合组织学图像信息增强空间转录组细胞聚类的分辨率[J]. 生物技术通报, 2024, 40(8): 39-46. |
| [6] | 高萌萌, 赵天宇, 焦馨悦, 林春晶, 关哲允, 丁孝羊, 孙妍妍, 张春宝. 大豆细胞质雄性不育系及其恢复系的比较转录组分析[J]. 生物技术通报, 2024, 40(7): 137-149. |
| [7] | 廖杨梅, 赵国春, 翁学煌, 贾黎明, 陈仲. 无患子雄性不育品种‘琦蕊’不同发育时期雄花转录组分析[J]. 生物技术通报, 2024, 40(7): 197-206. |
| [8] | 虞昕磊, 何结望, 林国平, 李金海, 王大爱, 袁跃斌, 刘圣高, 李志豪, 陶德欣. 夏冬两季发酵雪茄烟叶的代谢组差异分析[J]. 生物技术通报, 2024, 40(6): 260-270. |
| [9] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [10] | 白志元, 徐菲, 杨午, 王明贵, 杨玉花, 张海平, 张瑞军. 大豆细胞质雄性不育弱恢复型杂种F1育性转变的转录组分析[J]. 生物技术通报, 2024, 40(6): 134-142. |
| [11] | 吴迪, 游小凤, 郑亦铮, 林楠, 张燕燕, 魏艺聪. 草珊瑚中类胡萝卜素合成的内源激素调控机制分析[J]. 生物技术通报, 2024, 40(5): 203-214. |
| [12] | 郭纯, 宋桂梅, 闫艳, 邸鹏, 王英平. 西洋参bZIP基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2024, 40(4): 167-178. |
| [13] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [14] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [15] | 谢倩, 江来, 贺进, 刘玲玲, 丁明月, 陈清西. 不同鲜食品质橄榄果实转录组测序及酚类代谢途径相关调控基因挖掘[J]. 生物技术通报, 2024, 40(3): 215-228. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||