








生物技术通报 ›› 2024, Vol. 40 ›› Issue (3): 229-241.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0839
江林琪1( ), 赵佳莹1, 郑飞雄1, 姚馨怡1, 李效贤1, 俞振明1,2(
), 赵佳莹1, 郑飞雄1, 姚馨怡1, 李效贤1, 俞振明1,2( )
)
收稿日期:2023-08-28
出版日期:2024-03-26
发布日期:2024-04-08
通讯作者:
俞振明,男,博士,副研究员,研究方向:中药资源开发;E-mail: yuzhenming@zcmu.edu.cn作者简介:江林琪,女,研究方向:逆境与药用植物次生代谢;E-mail: 2978151322@qq.com
基金资助:
JIANG Lin-qi1( ), ZHAO Jia-ying1, ZHENG Fei-xiong1, YAO Xin-yi1, LI Xiao-xian1, YU Zhen-ming1,2(
), ZHAO Jia-ying1, ZHENG Fei-xiong1, YAO Xin-yi1, LI Xiao-xian1, YU Zhen-ming1,2( )
)
Received:2023-08-28
Published:2024-03-26
Online:2024-04-08
摘要:
【目的】 14-3-3蛋白,亦称通用调节因子(GRF),由多基因家族编码,在植物生长发育和逆境应答发挥关键的作用。鉴定铁皮石斛(Dendrobium officinale)GRF基因家族,为铁皮石斛GRF基因功能研究及遗传改良提供理论依据。【方法】 通过生物信息学的方法鉴定铁皮石斛14-3-3家族成员,分析其理化性质、染色体定位、系统进化发育、基因结构和启动子顺式作用元件等,同时通过荧光定量PCR技术检测它们在不同组织、低温处理及盐胁迫处理后的表达量。【结果】 铁皮石斛有17个GRF家族成员,分为ε类和非ε类亚族,不均匀地分布在7条染色体上,且存在7对串联复制基因。同一亚族成员基因结构、保守基序和蛋白质二级结构相类似。DoGRF家族基因的启动子区域存在大量激素和环境胁迫应答相关的调控元件。DoGRF家族基因在铁皮石斛各组织中均有表达,具有组织表达特异性,大多数基因在花器官中表达最高,其次是茎和根。同时,在低温处理、盐胁迫处理下呈现差异化表达,可能受到低温和盐胁迫的调控,特别是DoGRF2在铁皮石斛逆境应答过程中起着关键的作用。【结论】 在全基因组水平从铁皮石斛中鉴定出17个DoGRF家族成员,不同基因对非生物胁迫响应不一,并具有器官表达特异性。DoGRF2对低温和盐胁迫呈现正向响应。
江林琪, 赵佳莹, 郑飞雄, 姚馨怡, 李效贤, 俞振明. 铁皮石斛14-3-3基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(3): 229-241.
JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale[J]. Biotechnology Bulletin, 2024, 40(3): 229-241.
| 引物名称Primer name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| EF-1α | GCTTGAGAAGGAGCCCAAGT | CCAACAGCCACAGTTTGTCG |
| DoGRF1 | GAAGCTGCTGAACAGTCATTG | CCCAGTCTAATTGGGTGAGTAG |
| DoGRF2 | AAAGAAGAGAGTCGTGGGAATG | GTAGCAGAAGGAATGAGATGGG |
| DoGRF3 | GCTGGAATCCTTCGTTTGTTG | TACCGATGGTAATCGCCTTTC |
| DoGRF4 | CGGTGGTATCCTCTCTTTGTTAG | AGCAAGATAGCGGTGGTAATC |
| DoGRF5 | AGCATCACGTGAAGAGGATTAG | CAGCAGACGACGAAGGAATAA |
| DoGRF6 | GAGGATCGTGTGGCGATTAT | GTAGCAGAGGGAATCAGATGAG |
| DoGRF7 | CCATAGGTACCTTGCTGAGTTT | AGTGCCAATCCCAACCTTATAG |
| DoGRF8 | CGGAGATGAAGGTATCCAACAG | TTGCAGTGTCCTCTACCAATC |
| DoGRF9 | GAGATGCTGCAATCGCTAATTC | CCGCTACACTAAACGCAAATG |
| DoGRF10 | ATCCGGCTCTTGAAGGAATG | GGAGCCAGAACAGACAAAGT |
| DoGRF11 | GGATGCGGAAGTCGCTAAT | ATGCTCGGCCAGACTAAAC |
| DoGRF12 | CCTCATTTCCGTCGCATACA | GCCTTCTTCCTTCTGCTCAA |
| DoGRF13 | AAAGTATATGGCGGAGGTGAAG | AATTGCTCTTCGTCCTCCTTAG |
| DoGRF14 | CATCGAGCAGAAGGAGGAAA | GGTGTGAGTCGAGTAACCTAAG |
| DoGRF15 | GGCTGAGAGGTATGAGGAAATG | AGGCTACCGAGAGGAGATTT |
| DoGRF16 | GCGGAGGATATTGCTCTTGT | GGCGAGCGTTCTCTGATAAA |
| DoGRF17 | TGAGAGAGAGCAGCAAGTTTAC | CTACTGTCAGCTCCACATCTAATC |
表1 本实验所用的引物序列
Table 1 Primer sequences used in this study
| 引物名称Primer name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| EF-1α | GCTTGAGAAGGAGCCCAAGT | CCAACAGCCACAGTTTGTCG |
| DoGRF1 | GAAGCTGCTGAACAGTCATTG | CCCAGTCTAATTGGGTGAGTAG |
| DoGRF2 | AAAGAAGAGAGTCGTGGGAATG | GTAGCAGAAGGAATGAGATGGG |
| DoGRF3 | GCTGGAATCCTTCGTTTGTTG | TACCGATGGTAATCGCCTTTC |
| DoGRF4 | CGGTGGTATCCTCTCTTTGTTAG | AGCAAGATAGCGGTGGTAATC |
| DoGRF5 | AGCATCACGTGAAGAGGATTAG | CAGCAGACGACGAAGGAATAA |
| DoGRF6 | GAGGATCGTGTGGCGATTAT | GTAGCAGAGGGAATCAGATGAG |
| DoGRF7 | CCATAGGTACCTTGCTGAGTTT | AGTGCCAATCCCAACCTTATAG |
| DoGRF8 | CGGAGATGAAGGTATCCAACAG | TTGCAGTGTCCTCTACCAATC |
| DoGRF9 | GAGATGCTGCAATCGCTAATTC | CCGCTACACTAAACGCAAATG |
| DoGRF10 | ATCCGGCTCTTGAAGGAATG | GGAGCCAGAACAGACAAAGT |
| DoGRF11 | GGATGCGGAAGTCGCTAAT | ATGCTCGGCCAGACTAAAC |
| DoGRF12 | CCTCATTTCCGTCGCATACA | GCCTTCTTCCTTCTGCTCAA |
| DoGRF13 | AAAGTATATGGCGGAGGTGAAG | AATTGCTCTTCGTCCTCCTTAG |
| DoGRF14 | CATCGAGCAGAAGGAGGAAA | GGTGTGAGTCGAGTAACCTAAG |
| DoGRF15 | GGCTGAGAGGTATGAGGAAATG | AGGCTACCGAGAGGAGATTT |
| DoGRF16 | GCGGAGGATATTGCTCTTGT | GGCGAGCGTTCTCTGATAAA |
| DoGRF17 | TGAGAGAGAGCAGCAAGTTTAC | CTACTGTCAGCTCCACATCTAATC |
| 蛋白名称 Protein name | 氨基酸数Number of amino acids/aa | 分子量Molecular weight/kD | 等电点Isoelectric point | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水系数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| DoGRF1 | 248 | 28.24 | 4.85 | 43.69 | 86.98 | -0.444 | 细胞核Nucleus |
| DoGRF2 | 299 | 34.17 | 5.18 | 42.37 | 91.10 | -0.341 | 细胞核Nucleus |
| DoGRF3 | 265 | 29.77 | 4.96 | 41.61 | 91.40 | -0.283 | 细胞核Nucleus |
| DoGRF4 | 266 | 29.93 | 4.96 | 43.50 | 93.31 | -0.293 | 细胞核Nucleus |
| DoGRF5 | 267 | 30.00 | 4.83 | 39.17 | 81.24 | -0.537 | 细胞核Nucleus |
| DoGRF6 | 261 | 29.48 | 6.40 | 42.95 | 87.13 | -0.357 | 细胞核Nucleus |
| DoGRF7 | 250 | 28.25 | 4.99 | 43.68 | 86.04 | -0.358 | 细胞核Nucleus |
| DoGRF8 | 267 | 30.31 | 4.87 | 41.45 | 80.41 | -0.552 | 细胞核Nucleus |
| DoGRF9 | 260 | 29.48 | 4.74 | 47.93 | 91.23 | -0.483 | 细胞核Nucleus |
| DoGRF10 | 257 | 28.94 | 4.73 | 48.82 | 83.23 | -0.472 | 细胞核Nucleus |
| DoGRF11 | 257 | 29.10 | 4.82 | 49.74 | 86.65 | -0.503 | 细胞核Nucleus |
| DoGRF12 | 257 | 28.95 | 4.85 | 48.62 | 81.36 | -0.508 | 细胞核Nucleus |
| DoGRF13 | 254 | 28.57 | 5.94 | 51.58 | 90.39 | -0.259 | 细胞核Nucleus |
| DoGRF14 | 146 | 16.29 | 6.83 | 50.72 | 79.66 | -0.125 | 叶绿体Chloroplast |
| DoGRF15 | 278 | 30.57 | 9.05 | 45.76 | 94.21 | -0.042 | 细胞核Nucleus |
| DoGRF16 | 266 | 29.51 | 8.24 | 40.65 | 95.86 | -0.021 | 细胞核Nucleus |
| DoGRF17 | 246 | 27.61 | 9.03 | 39.78 | 94.47 | -0.128 | 叶绿体Chloroplast |
表2 铁皮石斛DoGRF蛋白的理化性质分析
Table 2 Physicochemical properties of DoGRF proteins in D. officinale
| 蛋白名称 Protein name | 氨基酸数Number of amino acids/aa | 分子量Molecular weight/kD | 等电点Isoelectric point | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 总平均亲水系数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| DoGRF1 | 248 | 28.24 | 4.85 | 43.69 | 86.98 | -0.444 | 细胞核Nucleus |
| DoGRF2 | 299 | 34.17 | 5.18 | 42.37 | 91.10 | -0.341 | 细胞核Nucleus |
| DoGRF3 | 265 | 29.77 | 4.96 | 41.61 | 91.40 | -0.283 | 细胞核Nucleus |
| DoGRF4 | 266 | 29.93 | 4.96 | 43.50 | 93.31 | -0.293 | 细胞核Nucleus |
| DoGRF5 | 267 | 30.00 | 4.83 | 39.17 | 81.24 | -0.537 | 细胞核Nucleus |
| DoGRF6 | 261 | 29.48 | 6.40 | 42.95 | 87.13 | -0.357 | 细胞核Nucleus |
| DoGRF7 | 250 | 28.25 | 4.99 | 43.68 | 86.04 | -0.358 | 细胞核Nucleus |
| DoGRF8 | 267 | 30.31 | 4.87 | 41.45 | 80.41 | -0.552 | 细胞核Nucleus |
| DoGRF9 | 260 | 29.48 | 4.74 | 47.93 | 91.23 | -0.483 | 细胞核Nucleus |
| DoGRF10 | 257 | 28.94 | 4.73 | 48.82 | 83.23 | -0.472 | 细胞核Nucleus |
| DoGRF11 | 257 | 29.10 | 4.82 | 49.74 | 86.65 | -0.503 | 细胞核Nucleus |
| DoGRF12 | 257 | 28.95 | 4.85 | 48.62 | 81.36 | -0.508 | 细胞核Nucleus |
| DoGRF13 | 254 | 28.57 | 5.94 | 51.58 | 90.39 | -0.259 | 细胞核Nucleus |
| DoGRF14 | 146 | 16.29 | 6.83 | 50.72 | 79.66 | -0.125 | 叶绿体Chloroplast |
| DoGRF15 | 278 | 30.57 | 9.05 | 45.76 | 94.21 | -0.042 | 细胞核Nucleus |
| DoGRF16 | 266 | 29.51 | 8.24 | 40.65 | 95.86 | -0.021 | 细胞核Nucleus |
| DoGRF17 | 246 | 27.61 | 9.03 | 39.78 | 94.47 | -0.128 | 叶绿体Chloroplast |

图3 铁皮石斛、水稻、拟南芥、大豆、蒺藜苜蓿、非洲菊GRF家族成员的系统发育树
Fig. 3 Phylogenetic tree of GRF family members in D. of-ficinale, O. sativa, A. thaliana, G. max, M. truncatula and G. hybrida
| 蛋白名称Protein name | α-螺旋 Alpha helix | 延伸链 Extended strand | β-折叠 Beta turn | 随机卷曲 Random coil |
|---|---|---|---|---|
| DoGRF1 | 52.42 | 12.54 | 5.41 | 29.63 |
| DoGRF2 | 62.54 | 11.04 | 3.34 | 23.08 |
| DoGRF3 | 49.75 | 14.39 | 5.30 | 30.56 |
| DoGRF4 | 52.31 | 14.58 | 6.25 | 26.85 |
| DoGRF5 | 59.20 | 11.66 | 3.37 | 25.77 |
| DoGRF6 | 67.43 | 6.51 | 3.07 | 22.99 |
| DoGRF7 | 52.96 | 12.81 | 5.17 | 29.06 |
| DoGRF8 | 50.84 | 10.60 | 5.06 | 33.49 |
| DoGRF9 | 51.75 | 11.05 | 4.85 | 32.35 |
| DoGRF10 | 52.48 | 15.45 | 2.04 | 30.03 |
| DoGRF11 | 52.06 | 11.60 | 4.38 | 31.96 |
| DoGRF12 | 49.77 | 10.42 | 5.09 | 34.72 |
| DoGRF13 | 44.19 | 16.28 | 5.04 | 34.50 |
| DoGRF14 | 34.91 | 15.98 | 10.65 | 38.46 |
| DoGRF15 | 46.38 | 13.62 | 8.09 | 31.91 |
| DoGRF16 | 45.36 | 12.500 | 5.85 | 36.29 |
| DoGRF17 | 43.03 | 13.75 | 6.97 | 36.25 |
表3 铁皮石斛DoGRF成员的蛋白二级结构分析
Table 3 Analysis of secondary structure of DoGRF proteins in D. officinale
| 蛋白名称Protein name | α-螺旋 Alpha helix | 延伸链 Extended strand | β-折叠 Beta turn | 随机卷曲 Random coil |
|---|---|---|---|---|
| DoGRF1 | 52.42 | 12.54 | 5.41 | 29.63 |
| DoGRF2 | 62.54 | 11.04 | 3.34 | 23.08 |
| DoGRF3 | 49.75 | 14.39 | 5.30 | 30.56 |
| DoGRF4 | 52.31 | 14.58 | 6.25 | 26.85 |
| DoGRF5 | 59.20 | 11.66 | 3.37 | 25.77 |
| DoGRF6 | 67.43 | 6.51 | 3.07 | 22.99 |
| DoGRF7 | 52.96 | 12.81 | 5.17 | 29.06 |
| DoGRF8 | 50.84 | 10.60 | 5.06 | 33.49 |
| DoGRF9 | 51.75 | 11.05 | 4.85 | 32.35 |
| DoGRF10 | 52.48 | 15.45 | 2.04 | 30.03 |
| DoGRF11 | 52.06 | 11.60 | 4.38 | 31.96 |
| DoGRF12 | 49.77 | 10.42 | 5.09 | 34.72 |
| DoGRF13 | 44.19 | 16.28 | 5.04 | 34.50 |
| DoGRF14 | 34.91 | 15.98 | 10.65 | 38.46 |
| DoGRF15 | 46.38 | 13.62 | 8.09 | 31.91 |
| DoGRF16 | 45.36 | 12.500 | 5.85 | 36.29 |
| DoGRF17 | 43.03 | 13.75 | 6.97 | 36.25 |
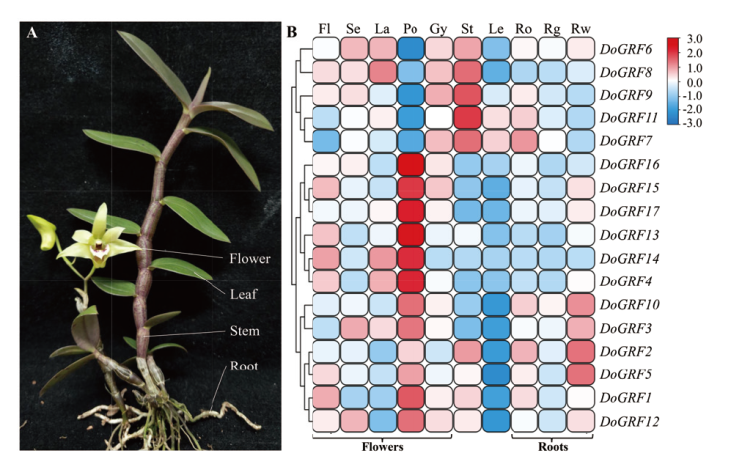
图6 铁皮石斛DoGRF家族基因的组织特异性表达分析 Fl:花蕾;Se:萼片;La:唇瓣;Po:花粉;Gy:合蕊柱;St:茎;Le:叶;Ro:根;Rg:绿色根尖;Rw:白色气生根
Fig. 6 Tissue specific expression analysis of DoGRF family genes in D. officinale Fl:Flower bud;Se:sepal;La:labellum;Po:pollinium;Gy:gynostemium;St:stem;Le:leaf;Ro:root;Rg:green root tip;Rw:white part root
| [1] |
Zhang HM, Zhu JH, Gong ZZ, et al. Abiotic stress responses in plants[J]. Nat Rev Genet, 2022, 23(2): 104-119.
doi: 10.1038/s41576-021-00413-0 |
| [2] |
严静, 蔡易熹, 陈燕兰, 等. 铁皮石斛茎、叶、花的活性成分及综合利用研究进展[J]. 食品与发酵工业, 2021, 47(17): 299-306.
doi: 10.13995/j.cnki.11-1802/ts.026336 |
| Yan J, Cai YX, Chen YL, et al. Research progress in active components and comprehensive utilization of stems, leaves and flowers of Dendrobium officinale[J]. Food Ferment Ind, 2021, 47(17): 299-306. | |
| [3] |
Yu ZM, Yang ZY, Teixeira da Silva JA, et al. Influence of low temperature on physiology and bioactivity of postharvest Dendrobium officinale stems[J]. Postharvest Biol Technol, 2019, 148: 97-106.
doi: 10.1016/j.postharvbio.2018.10.014 URL |
| [4] |
Niu ZT, Zhu F, Fan YJ, et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study[J]. Acta Pharm Sin B, 2021, 11(7): 2080-2092.
doi: 10.1016/j.apsb.2021.01.019 URL |
| [5] |
Zhao X, Li F, Li K. The 14-3-3 proteins: regulators of plant metabolism and stress responses[J]. Plant Biol, 2021, 23(4): 531-539.
doi: 10.1111/plb.v23.4 URL |
| [6] |
Ren JX, Zhang P, Dai YB, et al. Evolution of the 14-3-3 gene family in monocotyledons and dicotyledons and validation of MdGRF13 function in transgenic Arabidopsis thaliana[J]. Plant Cell Rep, 2023, 42(8): 1345-1364.
doi: 10.1007/s00299-023-03035-4 |
| [7] |
Huang Y, Wang WS, Yu H, et al. The role of 14-3-3 proteins in plant growth and response to abiotic stress[J]. Plant Cell Rep, 2022, 41(4): 833-852.
doi: 10.1007/s00299-021-02803-4 |
| [8] | 李芳, 滕建晒, 陈宣钦. 14-3-3蛋白参与植物应答非生物胁迫的研究进展[J]. 植物科学学报, 2018, 36(3): 459-469. |
| Li F, Teng JS, Chen XQ. Research progress on the 14-3-3 protein involved in plant responses to abiotic stress[J]. Plant Sci J, 2018, 36(3): 459-469. | |
| [9] | 桑婷, 颜秀娟, 裴红霞, 等. 辣椒14-3-3蛋白基因家族全基因组鉴定与表达特征分析[J]. 分子植物育种, 2021, 19(16): 5268-5278. |
| Sang T, Yan XJ, Pei HX, et al. Genome-wide identification and expression characteristics analysis of 14-3-3 gene family in pepper(Capsicum annuum)[J]. Mol Plant Breed, 2021, 19(16): 5268-5278. | |
| [10] |
He FY, Duan SG, Jian YQ, et al. Genome-wide identification and gene expression analysis of the 14-3-3 gene family in potato(Solanum tuberosum L.)[J]. BMC Genomics, 2022, 23(1): 811.
doi: 10.1186/s12864-022-09037-y |
| [11] |
Xu MY, Hu ZY, Lai W, et al. Comprehensive analysis of 14-3-3 family genes and their responses to cold and drought stress in cucumber[J]. Funct Plant Biol, 2021, 48(12): 1264-1276.
doi: 10.1071/FP21022 pmid: 34635203 |
| [12] |
Chen F, Li Q, Sun LX, et al. The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress[J]. DNA Res, 2006, 13(2): 53-63.
doi: 10.1093/dnares/dsl001 URL |
| [13] |
Pan RR, Wang YJ, An FF, et al. Genome-wide identification and characterization of 14-3-3 gene family related to negative regulation of starch accumulation in storage root of Manihot esculenta[J]. Front Plant Sci, 2023, 14: 1184903.
doi: 10.3389/fpls.2023.1184903 URL |
| [14] |
Zhang ZB, Wang XK, Wang S, et al. Expansion and diversification of the 14-3-3 gene family in Camellia sinensis[J]. J Mol Evol, 2022, 90(3-4): 296-306.
doi: 10.1007/s00239-022-10060-6 |
| [15] |
Liu Q, Zhang SH, Liu B. 14-3-3 proteins: Macro-regulators with great potential for improving abiotic stress tolerance in plants[J]. Biochem Biophys Res Commun, 2016, 477(1): 9-13.
doi: 10.1016/j.bbrc.2016.05.120 URL |
| [16] |
Camoni L, Visconti S, Aducci P, et al. 14-3-3 proteins in plant hormone signaling: doing several things at once[J]. Front Plant Sci, 2018, 9: 297.
doi: 10.3389/fpls.2018.00297 pmid: 29593761 |
| [17] |
Yang ZJ, Wang CW, Xue Y, et al. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance[J]. Nat Commun, 2019, 10(1): 1199.
doi: 10.1038/s41467-019-09181-2 pmid: 30867421 |
| [18] |
Yu ZM, Zhang GH, Teixeira da Silva JA, et al. Genome-wide identification and analysis of DNA methyltransferase and demethylase gene families in Dendrobium officinale reveal their potential functions in polysaccharide accumulation[J]. BMC Plant Biol, 2021, 21(1): 21.
doi: 10.1186/s12870-020-02811-8 |
| [19] |
Zhang MZ, Liu N, Teixeira da Silva JA, et al. Physiological and transcriptomic analysis uncovers salinity stress mechanisms in a facultative crassulacean acid metabolism plant Dendrobium officinale[J]. Front Plant Sci, 2022, 13: 1028245.
doi: 10.3389/fpls.2022.1028245 URL |
| [20] |
Lu SN, Wang JY, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020[J]. Nucleic Acids Res, 2020, 48(D1): D265-D268.
doi: 10.1093/nar/gkz991 URL |
| [21] |
Potter SC, Luciani A, Eddy SR, et al. HMMER web server: 2018 update[J]. Nucleic Acids Res, 2018, 46(W1): W200-W204.
doi: 10.1093/nar/gky448 URL |
| [22] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [23] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [24] |
Zhang GQ, Liu KW, Li Z, et al. The Apostasia genome and the evolution of orchids[J]. Nature, 2017, 549(7672): 379-383.
doi: 10.1038/nature23897 URL |
| [25] |
Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown[J]. Nat Protoc, 2016, 11(9): 1650-1667.
doi: 10.1038/nprot.2016.095 pmid: 27560171 |
| [26] | 俞振明, 赵聪慧, 张明泽, 等. 益智不同组织多糖含量及其生物合成途径分析[J]. 热带亚热带植物学报, 2021, 29(6): 669-677. |
| Yu ZM, Zhao CH, Zhang MZ, et al. Analysis of polysaccharide content and biosynthesis pathway in different tissues of Alpinia oxyphylla[J]. J Trop Subtrop Bot, 2021, 29(6): 669-677. | |
| [27] |
Yu ZM, Zhang GH, Teixeira da Silva JA, et al. The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development[J]. Plant Sci, 2021, 309: 110952.
doi: 10.1016/j.plantsci.2021.110952 URL |
| [28] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [29] |
Liang YF, Ma F, Zhang RL, et al. Genome-wide identification and characterization of tomato 14-3-3(SlTFT)genes and functional analysis of SlTFT6 under heat stress[J]. Physiol Plant, 2023, 175(2): e13888.
doi: 10.1111/ppl.v175.2 URL |
| [30] |
Zuo XY, Wang SX, Xiang W, et al. Genome-wide identification of the 14-3-3 gene family and its participation in floral transition by interacting with TFL1/FT in apple[J]. BMC Genomics, 2021, 22(1): 41.
doi: 10.1186/s12864-020-07330-2 |
| [31] |
Liu ZY, Jia YX, Ding YL, et al. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response[J]. Mol Cell, 2017, 66(1): 117-128.
doi: S1097-2765(17)30131-4 pmid: 28344081 |
| [32] |
Wang NN, Shi YY, Jiang Q, et al. A 14-3-3 protein positively regulates rice salt tolerance by stabilizing phospholipase C1[J]. Plant Cell Environ, 2023, 46(4): 1232-1248.
doi: 10.1111/pce.14520 URL |
| [1] | 张玉, 石磊, 巩檑, 聂峰杰, 杨江伟, 刘璇, 杨文静, 张国辉, 颉瑞霞, 张丽. 马铃薯WOX基因家族的鉴定及在离体再生和非生物胁迫中的表达分析[J]. 生物技术通报, 2024, 40(3): 170-180. |
| [2] | 吴星星, 洪海波, 甘志承, 李瑞宁, 黄先忠. 辣椒CaPI的克隆与功能分析[J]. 生物技术通报, 2024, 40(3): 193-201. |
| [3] | 周宏丹, 罗晓萍, 涂米雪, 李忠光. 植物褪黑素:植物应答非生物胁迫的新兴信号分子[J]. 生物技术通报, 2024, 40(3): 41-51. |
| [4] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析[J]. 生物技术通报, 2024, 40(2): 130-145. |
| [5] | 辛奇, 李压凡, 尹铮, 张晓丹, 陈霆, 刘晓华. 甘蔗CBL-CIPK基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(2): 197-211. |
| [6] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [7] | 张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206. |
| [8] | 吴圳, 张明英, 闫锋, 李依民, 高静, 颜永刚, 张岗. 掌叶大黄(Rheum palmatum L.)WRKY基因家族鉴定与分析[J]. 生物技术通报, 2024, 40(1): 250-261. |
| [9] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [10] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [11] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [12] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [13] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [14] | 刘奎, 李兴芬, 杨沛欣, 仲昭晨, 曹一博, 张凌云. 青杄转录共激活因子PwMBF1c的功能研究与验证[J]. 生物技术通报, 2023, 39(5): 205-216. |
| [15] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||