








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 255-263.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1074
钟匀1( ), 林春1,2, 刘正杰1,2, 董陈文华1,2, 毛自朝1,2(
), 林春1,2, 刘正杰1,2, 董陈文华1,2, 毛自朝1,2( ), 李兴玉1,2,3(
), 李兴玉1,2,3( )
)
收稿日期:2023-11-14
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
李兴玉,男,教授,硕士生导师,研究方向:天然产物及生物防治;E-mail: lixingyu@ynau.edu.cn;作者简介:钟匀,男,硕士研究生,研究方向:药用植物合成生物学;E-mail: 1292726737@qq.com
基金资助:
ZHONG Yun1( ), LIN Chun1,2, LIU Zheng-jie1,2, DONG Chen-wen-hua1,2, MAO Zi-chao1,2(
), LIN Chun1,2, LIU Zheng-jie1,2, DONG Chen-wen-hua1,2, MAO Zi-chao1,2( ), LI Xing-yu1,2,3(
), LI Xing-yu1,2,3( )
)
Received:2023-11-14
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】克隆芦笋(Asparagus officinalis)甾醇糖基转移酶基因AoSGT1,分析其潜在催化活性,为解析芦笋皂苷合成途径及代谢调控机制提供科学依据。【方法】基于芦笋转录组数据设计特异性引物,扩增AoSGT1基因的完整开放阅读框(open reading frame, ORF),经测序验证获得目标基因序列并进行生物信息学分析;实时荧光定量PCR(quantitative real-time PCR, RT-qPCR)测定各组织基因表达量;构建pGEX-4T-3-AoSGT1原核表达载体,并转入大肠杆菌(Escherichia coli)BL21(De3),随后诱导实现重组蛋白表达。【结果】AoSGT1长1 800 bp,编码599个氨基酸,其相对分子质量为66.72 kD,属于亲水性蛋白,无跨膜域和信号肽。系统发育结果表明,AoSGT1与盾叶薯蓣(Dioscorea zingiberensis)Dz3GT2有较高同源性,同属UGT80B1亚家族。多序列比对揭示了该蛋白序列包含甾醇糖基转移酶保守结构域PSBD Box和PSPG Box,预示其具有对甾体化合物3β-OH位点潜在糖基化活性。RT-qPCR结果显示,AoSGT1在芦笋根中高表达,而在茎和花中低表达。此外,SDS-PAGE结果表明,目标蛋白在大肠杆菌中以可溶性形式高效表达,其大小与预测值相符。【结论】成功克隆AoSGT1基因,并确认其在芦笋中呈现组织特异性表达,推测其可能参与芦笋甾体皂苷生物合成。同时,成功在大肠杆菌中实现了目标蛋白的异源表达。
钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263.
ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes[J]. Biotechnology Bulletin, 2024, 40(4): 255-263.
| 引物Primer | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| UAoSGT1 | F:TCTTGATGCGATTACAGCGG R:GAGATGGCTACAAGGTGAAAGG | 基因克隆 Gene clone |
| PGEX-AoSGT1 | F:GTGGATCCCC GAATTCCATGAGGAGCGGAGATTTCGAAG R:GGCCGCTCGA GTCGACCTACAAGGTGAAAGGAAGACAACACC | 原核表达 Prokaryotic expression |
| qRT-AoSGT1 | F:ATCCTTTGGGTGGTGATCCG R:AGAGCTTCAGCAACATGCGT | 实时荧光定量PCR RT-qPCR |
| EF1A | F:CTGGCCAGGGTGGTTCATGAT R:TAAGTCTGTTGAGATGCACC | 内参基因 Reference gene |
表1 本研究涉及PCR引物序列
Table 1 PCR primer sequences used in this study
| 引物Primer | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| UAoSGT1 | F:TCTTGATGCGATTACAGCGG R:GAGATGGCTACAAGGTGAAAGG | 基因克隆 Gene clone |
| PGEX-AoSGT1 | F:GTGGATCCCC GAATTCCATGAGGAGCGGAGATTTCGAAG R:GGCCGCTCGA GTCGACCTACAAGGTGAAAGGAAGACAACACC | 原核表达 Prokaryotic expression |
| qRT-AoSGT1 | F:ATCCTTTGGGTGGTGATCCG R:AGAGCTTCAGCAACATGCGT | 实时荧光定量PCR RT-qPCR |
| EF1A | F:CTGGCCAGGGTGGTTCATGAT R:TAAGTCTGTTGAGATGCACC | 内参基因 Reference gene |
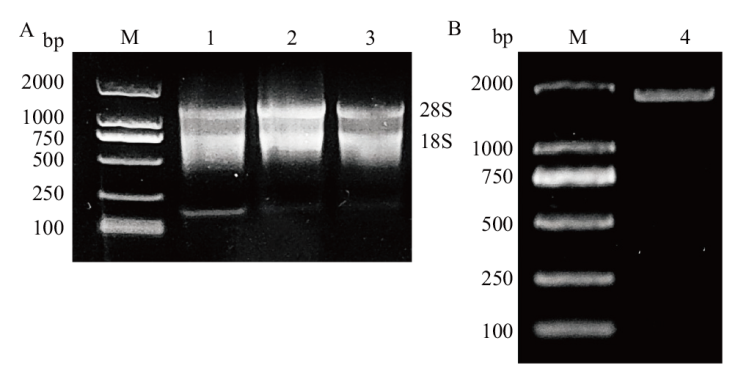
图1 芦笋RNA及AOSGT1基因扩增产物凝胶电泳 A:芦笋各组织总RNA琼脂糖凝胶电泳;B:AoSGT1基因扩增;M:DNA marker 2000;1:根;2:茎;3:花;4:AoSGT1
Fig. 1 Agarose gel electrophoresis of asparagus RNA and AOSGT1 gene amplification products A: Agarose gel electrophoresis of total RNA from various tissues of asparagus. B: Amplification of the AoSGT1 gene. M: DNA marker 2000. 1: Root. 2: Stem. 3: Flower. 4: AoSGT1
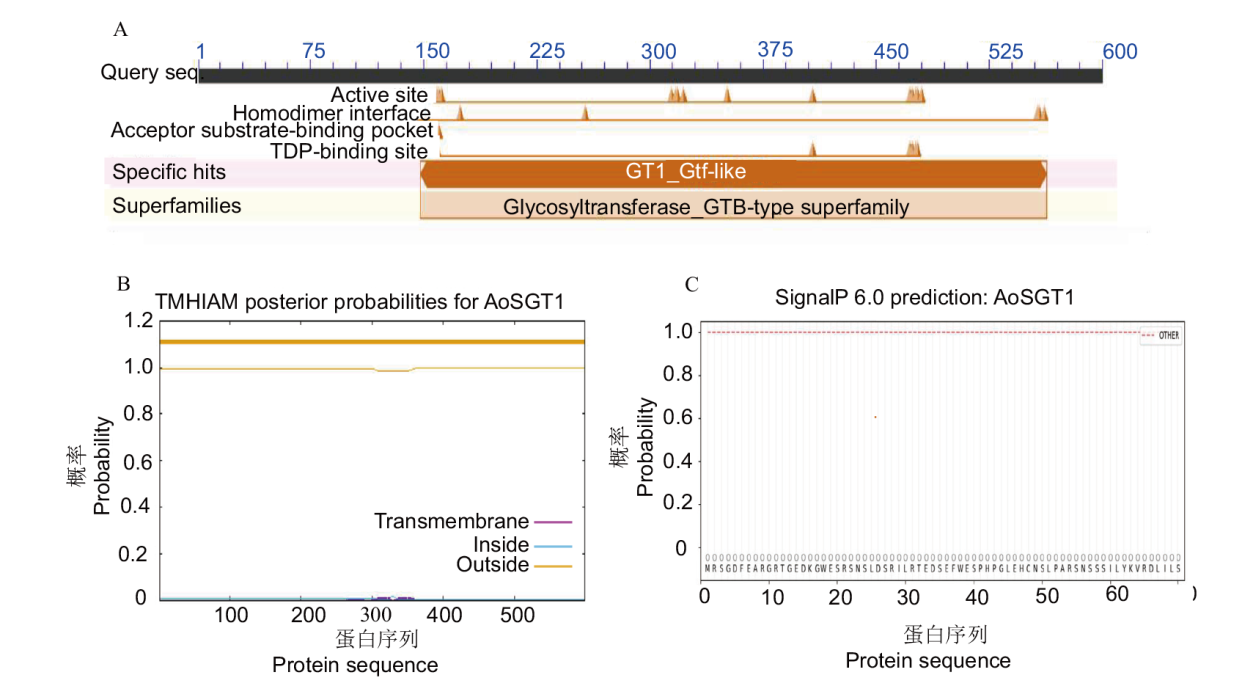
图2 AoSGT1的生物信息学分析 A:NCBI保守结构域预测;B:跨膜域预测;C:信号肽分析
Fig. 2 Bioinformatics analysis of AoSGT1 A: NCBI conservative structural domain prediction. B: Transmembrane domain prediction. C: Signal peptide analysis
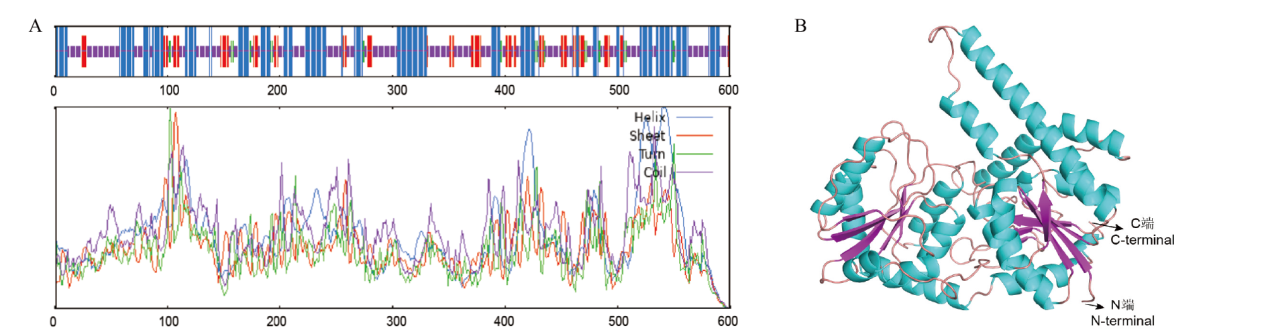
图3 芦笋AoSGT1蛋白的结构特征 A:AoSGT1二级结构(蓝:α-螺旋;绿:β-转角;橙:无规则卷曲;紫:无二级结构);B:AoSGT1三维结构预测(蓝:螺旋;粉红:折叠;橙:环)
Fig. 3 Structural characteristics of asparagus AoSGT1 protein A: AoSGT1 secondary structure(Blue: α- Spiral; green: β- Corner; orange: irregular curl; purple: no secondary structure). B: AoSGT1 3D structure prediction(Blue: helix; pink: sheet; orange: loop)
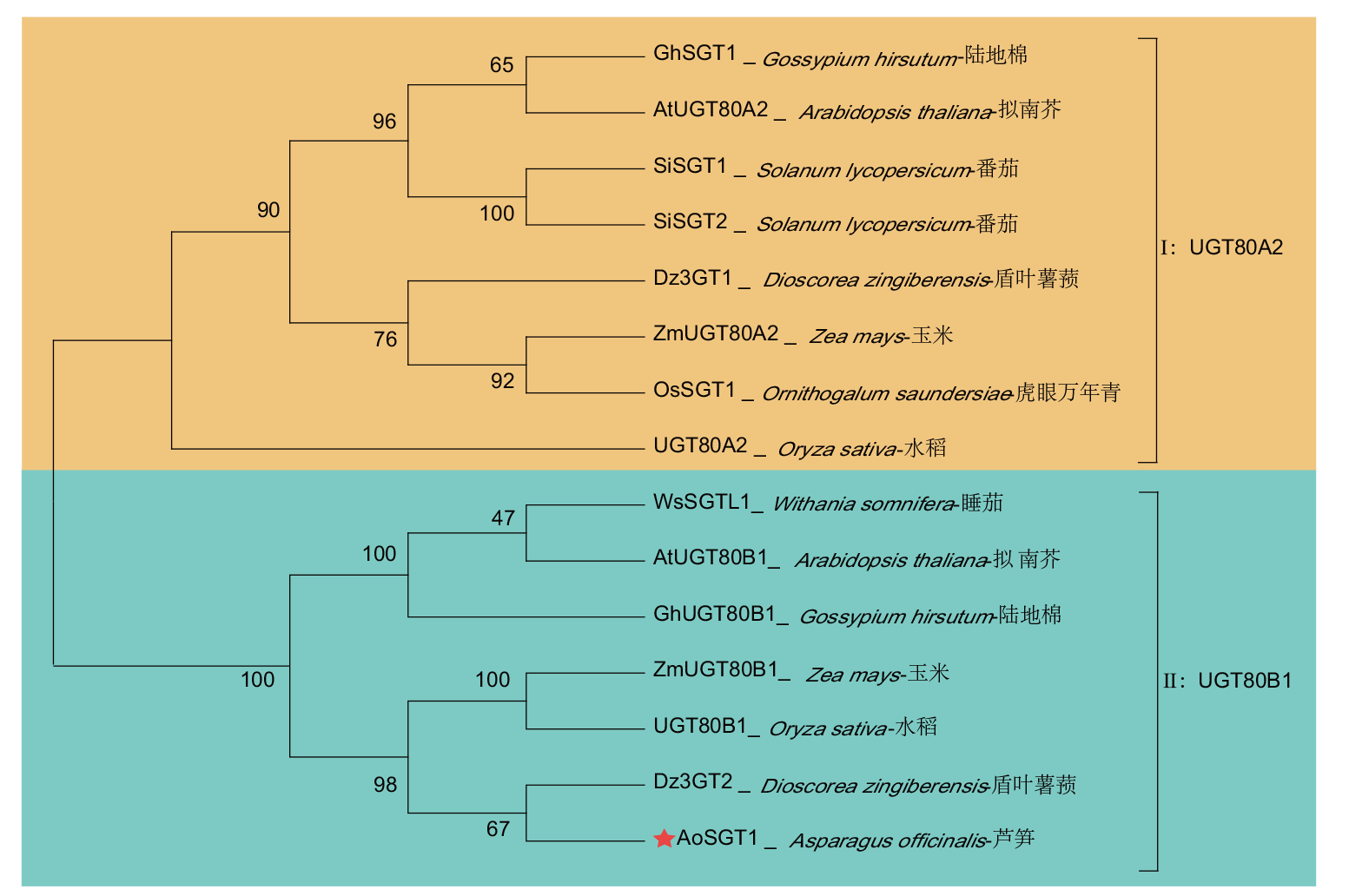
图4 芦笋AoSGT1与其他植物SGTs的系统进化树 GhSGT1: AHX00584; AtUGT80A2: OAP05643.1; SiSGT1: ATO74554.1; SiSGT2: ATO74555.1; Dz3GT1: AVI57699; ZmUGT80A2: PWZ44786.1; OsSGT1: AWW17242; UGT80A2: XP_015647949.1; WsSGTL1: ABC96116; AtUGT80B1: OAP13743.1; GhUGT80B1: NP: 001314026.1; ZmUGT80B1: PWZ33516.1; OsUGT80B1: XP_015622442.1; Dz3GT2: AVI57700
Fig. 4 Phylogenetic tree of asparagus AoSGT1 and SGTs from other plants
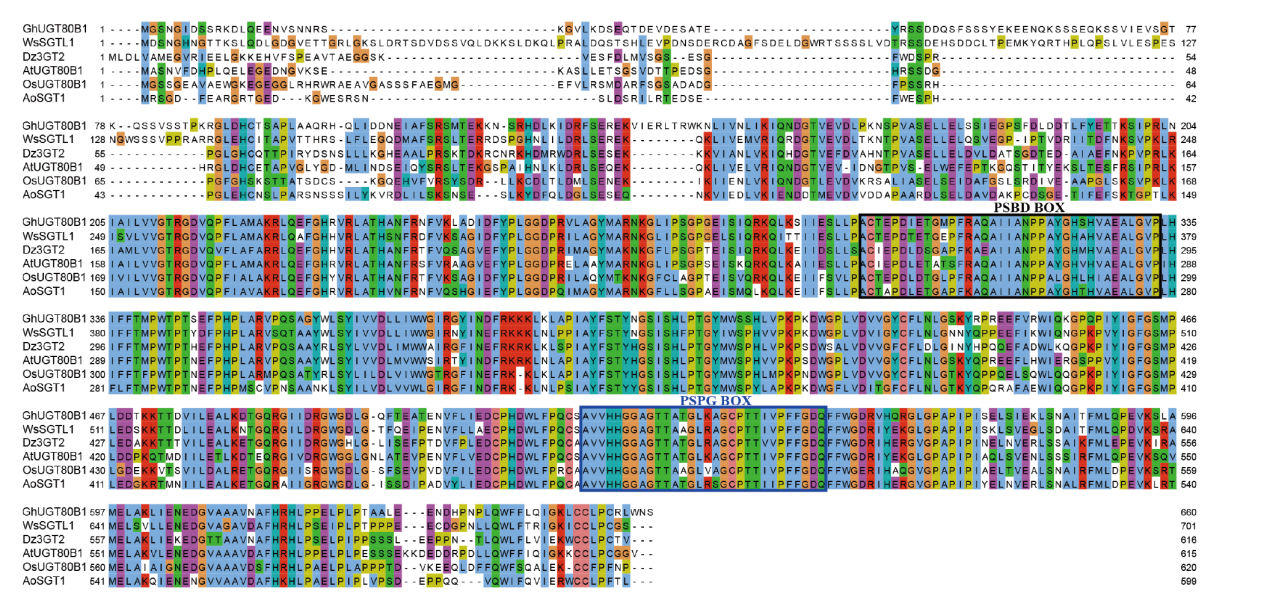
图5 芦笋AoSGT1多序列比对分析 黑色框区域:PSBD Box结构域;蓝色框区域:PSPG Box结构域
Fig. 5 Multiple sequence alignment analysis of asparagus AoSGT1 Black box area: PSBD Box structural domain; blue box area: PSPG Box structure domain
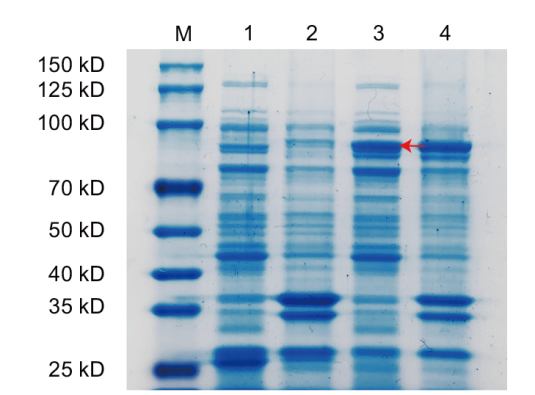
图7 AoSGT1原核表达的 SDS-PAGE凝胶电泳 M:蛋白marker;1:空载上清液;2:空载沉淀;3:AoSGT1上清液;4:AoSGT1沉淀
Fig. 7 SDS-PAGE gel electrophoresis of prokaryotic expression of AoSGT1 M: Protein marker. 1: Empty supernatant. 2: Empty sediment. 3: AoSGT1 supernatant. 4: AoSGT1 sediment
| [1] | 皇秋秋, 杜加欢, 尹俊玉, 等. 芦笋甾体皂苷合成与调控的研究进展[J]. 植物生理学报, 2020, 56(7): 1395-1407. |
| Huang QQ, Du JH, Yin JY, et al. Advances in biosynthesis and regulation of saponins from Asparagus officinalis[J]. Plant Physiol J, 2020, 56(7): 1395-1407. | |
| [2] |
Yu Q, Fan LP. Improving the bioactive ingredients and functions of asparagus from efficient to emerging processing technologies: a review[J]. Food Chem, 2021, 358: 129903.
doi: 10.1016/j.foodchem.2021.129903 URL |
| [3] |
Zhang HX, Birch J, Pei JJ, et al. Phytochemical compounds and biological activity in Asparagus roots: a review[J]. Int J Food Sci Tech, 2019, 54(4): 966-977.
doi: 10.1111/ijfs.2019.54.issue-4 URL |
| [4] | Li Y, Yang H, Li ZH, et al. Advances in the biosynthesis and molecular evolution of steroidal saponins in plants[J]. Int J Mol Sci, 2023, 24(3): 2620. |
| [5] |
Moreau RA, Nyström L, Whitaker BD, et al. Phytosterols and their derivatives: structural diversity, distribution, metabolism, analysis, and health-promoting uses[J]. Prog Lipid Res, 2018, 70: 35-61.
doi: S0163-7827(17)30062-0 pmid: 29627611 |
| [6] | Anwar Z, Hussain F. Steroidal saponins: an overview of medicinal uses[J]. Int J Chem & Biochem Sci, 2017,11: 20-24. |
| [7] |
Zhou C, Yang YH, Tian JY, et al. 22R- but not 22S-hydroxycholesterol is recruited for diosgenin biosynthesis[J]. Plant J, 2022, 109(4): 940-951.
doi: 10.1111/tpj.v109.4 URL |
| [8] |
Mohammadi M, Mashayekh T, Rashidi-Monfared S, et al. New insights into diosgenin biosynthesis pathway and its regulation in Trigonella foenum-graecum L[J]. Phytochem Anal, 2020, 31(2): 229-241.
doi: 10.1002/pca.v31.2 URL |
| [9] |
Yin Y, Gao LH, Zhang XN, et al. A cytochrome P450 monooxygenase responsible for the C-22 hydroxylation step in the Paris polyphylla steroidal saponin biosynthesis pathway[J]. Phytochemistry, 2018, 156: 116-123.
doi: 10.1016/j.phytochem.2018.09.005 URL |
| [10] | 秦晶晶, 孙春玉, 张美萍, 等. 植物UDP-糖基转移酶分类、功能以及进化[J]. 基因组学与应用生物学, 2018, 37(1): 440-450. |
| Qin JJ, Sun CY, Zhang MP, et al. Classification, function and evolution of plant UDP-glycosyltransferase[J]. Genom Appl Biol, 2018, 37(1): 440-450. | |
| [11] | Cantarel BL, Coutinho PM, Rancurel C, et al. The Carbohydrate-Active EnZymes database(CAZy): an expert resource for glycogenomics[J]. Nucleic Acids Res, 2009, 37(Database issue): D233-D238. |
| [12] |
Stucky DF, Arpin JC, Schrick K. Functional diversification of two UGT80 enzymes required for steryl glucoside synthesis in Arabidopsis[J]. J Exp Bot, 2015, 66(1): 189-201.
doi: 10.1093/jxb/eru410 URL |
| [13] | Tiwari P, Sangwan RS, Asha, et al. Molecular cloning and biochemical characterization of a recombinant sterol 3-O-glucosyltransferase from Gymnema sylvestre R.Br. catalyzing biosynthesis of steryl glucosides[J]. Biomed Res Int, 2014, 2014: 934351. |
| [14] |
Gao JH, Xu YH, Hua CK, et al. Molecular cloning and functional characterization of a sterol 3- O-glucosyltransferase involved in biosynthesis of steroidal saponins in Trigonella foenum-graecum[J]. Front Plant Sci, 2021, 12: 809579.
doi: 10.3389/fpls.2021.809579 URL |
| [15] | Li J, Liang Q, Li CF, et al. Comparative transcriptome analysis identifies putative genes involved in dioscin biosynthesis in Dioscorea zingiberensis[J]. Molecules, 2018, 23(2): 454. |
| [16] |
Song W, Zhang CC, Wu JL, et al. Characterization of three Paris polyphylla glycosyltransferases from different UGT families for steroid functionalization[J]. ACS Synth Biol, 2022, 11(4): 1669-1680.
doi: 10.1021/acssynbio.2c00103 pmid: 35286065 |
| [17] | Harkess A, Zhou JS, Xu CY, et al. The asparagus genome sheds light on the origin and evolution of a young Y chromosome[J]. Nat Commun, 2017, 8(1): 1279. |
| [18] | Cheng Q, Zeng LQ, Wen H, et al. Steroidal saponin profiles and their key genes for synthesis and regulation in Asparagus officinalis L. by joint analysis of metabolomics and transcriptomics[J]. BMC Plant Biol, 2023, 23(1): 207. |
| [19] |
Chen LQ, Zhang Y, Feng Y. Structural dissection of sterol glycosyltransferase UGT51 from Saccharomyces cerevisiae for substrate specificity[J]. J Struct Biol, 2018, 204(3): 371-379.
doi: 10.1016/j.jsb.2018.11.001 URL |
| [20] |
Upadhyay S, Jeena GS, et al. Shikha, Recent advances in steroidal saponins biosynthesis and in vitro production[J]. Planta, 2018, 248(3): 519-544.
doi: 10.1007/s00425-018-2911-0 pmid: 29748819 |
| [21] |
Liu M, Kong JQ. The enzymatic biosynthesis of acylated steroidal glycosides and their cytotoxic activity[J]. Acta Pharm Sin B, 2018, 8(6): 981-994.
doi: 10.1016/j.apsb.2018.04.006 pmid: 30505666 |
| [22] |
Zhang M, Li FD, Li K, et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra[J]. J Am Chem Soc, 2020, 142(7): 3506-3512.
doi: 10.1021/jacs.9b12211 pmid: 31986016 |
| [23] |
Yang M, Fehl C, Lees KV, et al. Functional and informatics analysis enables glycosyltransferase activity prediction[J]. Nat Chem Biol, 2018, 14(12): 1109-1117.
doi: 10.1038/s41589-018-0154-9 pmid: 30420693 |
| [24] | Zhao XY, Zheng L, Si JJ, et al. Immunocytochemical localization of saikosaponin-d in vegetative organs of Bupleurum scorzonerifolium Willd[J]. Bot Stud, 2013, 54(1): 32. |
| [25] | Ye T, Song W, Zhang JJ, et al. Identification and functional characterization of DzS3GT, a cytoplasmic glycosyltransferase catalyzing biosynthesis of diosgenin 3-O-glucoside in Dioscorea zingiberensis[J]. Plant Cell Tissue Organ Cult PCTOC, 2017, 129(3): 399-410. |
| [26] |
Jaiswal Y, Liang ZT, Ho A, et al. A comparative tissue-specific metabolite analysis and determination of protodioscin content in Asparagus species used in traditional Chinese medicine and Ayurveda by use of laser microdissection, UHPLC-QTOF/MS and LC-MS/MS[J]. Phytochem Anal, 2014, 25(6): 514-528.
doi: 10.1002/pca.2522 URL |
| [27] | Yi TG, Yeoung YR, Choi IY, et al. Transcriptome analysis of Asparagus officinalis reveals genes involved in the biosynthesis of rutin and protodioscin[J]. PLoS One, 2019, 14(7): e0219973. |
| [28] |
Kohara A, Nakajima C, Hashimoto K, et al. A novel glucosyltransferase involved in steroid saponin biosynthesis in Solanum aculeatissimum[J]. Plant Mol Biol, 2005, 57(2): 225-239.
doi: 10.1007/s11103-004-7204-2 URL |
| [29] | 张磊, 唐永凯, 李红霞, 等. 促进原核表达蛋白可溶性的研究进展[J]. 中国生物工程杂志, 2021, 41(S1): 138-149. |
| Zhang L, Tang YK, Li HX, et al. Advances in promoting solubility of prokaryotic expressed proteins[J]. China Biotechnol, 2021, 41(S1): 138-149. | |
| [30] |
Hayat SMG, Farahani N, Golichenari B, et al. Recombinant protein expression in Escherichia coli(E. coli): what we need to know[J]. Curr Pharm Des, 2018, 24(6): 718-725.
doi: 10.2174/1381612824666180131121940 URL |
| [1] | 杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129. |
| [2] | 郭纯, 宋桂梅, 闫艳, 邸鹏, 王英平. 西洋参bZIP基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2024, 40(4): 167-178. |
| [3] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [4] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [5] | 谢倩, 江来, 贺进, 刘玲玲, 丁明月, 陈清西. 不同鲜食品质橄榄果实转录组测序及酚类代谢途径相关调控基因挖掘[J]. 生物技术通报, 2024, 40(3): 215-228. |
| [6] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [7] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [8] | 朱毅, 柳唐镜, 宫国义, 张洁, 王晋芳, 张海英. 西瓜ClPP2C3克隆及表达分析[J]. 生物技术通报, 2024, 40(1): 243-249. |
| [9] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [10] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [11] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [12] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [13] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [14] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [15] | 梅欢, 李玥, 刘可蒙, 刘吉华. 小檗碱桥酶高效原核表达及生物合成l-SLR的研究[J]. 生物技术通报, 2023, 39(7): 277-287. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||