








生物技术通报 ›› 2025, Vol. 41 ›› Issue (8): 220-233.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0057
• 研究报告 • 上一篇
腊贵晓1,2( ), 赵玉龙1,2, 代丹丹1,2, 余永亮1,2, 郭红霞1,2, 史贵霞1,2, 贾慧1, 杨铁钢1,2(
), 赵玉龙1,2, 代丹丹1,2, 余永亮1,2, 郭红霞1,2, 史贵霞1,2, 贾慧1, 杨铁钢1,2( )
)
收稿日期:2025-01-14
出版日期:2025-08-26
发布日期:2025-08-14
通讯作者:
杨铁钢,男,博士,研究员,研究方向 :中药材生态栽培育种;E-mail: ytgha@163.com作者简介:腊贵晓,男,博士,副研究员,研究方向 :中药材生态栽培;E-mail: zju-l@163.com
基金资助:
LA Gui-xiao1,2( ), ZHAO Yu-long1,2, DAI Dan-dan1,2, YU Yong-liang1,2, GUO Hong-xia1,2, SHI Gui-xia1,2, JIA Hui1, YANG Tie-gang1,2(
), ZHAO Yu-long1,2, DAI Dan-dan1,2, YU Yong-liang1,2, GUO Hong-xia1,2, SHI Gui-xia1,2, JIA Hui1, YANG Tie-gang1,2( )
)
Received:2025-01-14
Published:2025-08-26
Online:2025-08-14
摘要:
目的 对红花质膜H+-ATPase(CtPMA)基因家族进行全基因组鉴定与分析,为研究该基因家族及其在响应低氮低磷胁迫中的功能奠定基础。 方法 利用生物信息学方法对CtPMA基因家族成员进行鉴定和系统的分析;利用实时荧光定量PCR(RT-qPCR)分析其组织表达特异性及在低氮低磷胁迫下的响应。 结果 在红花中共鉴定到10个CtPMAs(命名为CtPMA1-CtPMA10),其编码蛋白质的氨基酸数为785-958 aa,相对分子质量为85.23-105.52 kD,等电点为5.26-7.91,亚细胞位点预测显示全部位于细胞膜;都含有Cation_ATPase_N结构域、E1_E2 ATPase结构域和HAD-superfamily hydrolase subfamily ⅢA结构域;顺式作用元件预测显示CtPMA基因家族内广泛存在与生长发育、激素和胁迫相关的调控元件;基因共线性显示全基因组复制和片段复制在CtPMA基因家族进化中起到了重要作用。RT-qPCR结果表明,在根中高表达的CtPMA1和CtPMA7的表达量显著受到低氮低磷胁迫的诱导,说明CtPMA1和CtPMA7可能与红花根部吸收氮磷的功能密切相关。 结论 红花全基因组中共鉴定到10个CtPMA基因,其进化相对保守,在不同的组织部位具有不同的表达模式。当红花遭受低氮低磷胁迫时在根部高表达的CtPMA1和CtPMA7的转录水平显著被诱导,暗示其通过诱导自身转录水平以提高对氮磷的吸收和利用效率,用来抵御营养等逆境胁迫。
腊贵晓, 赵玉龙, 代丹丹, 余永亮, 郭红霞, 史贵霞, 贾慧, 杨铁钢. 红花质膜H+-ATPase基因家族成员鉴定及响应低氮低磷胁迫的表达分析[J]. 生物技术通报, 2025, 41(8): 220-233.
LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress[J]. Biotechnology Bulletin, 2025, 41(8): 220-233.
| 基因 Gene | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') | 长度Length (bp) |
|---|---|---|---|
| CtPMA1 | TTGACCAATCTGCCCTCA | CCTTCTGGAAATGCCCAA | 189 |
| CtPMA2 | CCACCAGATTGGCAAGACT | GGCATCAGCAGGAACGAT | 231 |
| CtPMA3 | AGATGCCGCCGTGTTAGT | GTCAAAGCCGACTGGTCAA | 117 |
| CtPMA4 | AGGGATGGGAGATGGTCTG | GCTTCTTGGTGACTGGTAAGG | 169 |
| CtPMA5 | TGACGACTGATGATGGTAACC | CAATCTGGAGGCTTTCCC | 181 |
| CtPMA6 | GCAGCCATTCTCGTTCCA | AGGGTGCTTTGTCACAGGA | 144 |
| CtPMA7 | GGAATCCTCTCTCGTGGGT | GCAAGCATCTTCTTCGCTC | 242 |
| CtPMA8 | GGTGCTTCTTCTCATAAACTCG | ACGATGTCTCCTGGAACCA | 162 |
| CtPMA9 | TGAAGGCATTGATAACCTGC | ACACCAGCGGCAAATACC | 246 |
| CtPMA10 | GGCGAGTCTATTCCAGTAACC | GATTGGTGTTCTCCACAAGG | 148 |
| 18S | ACACGGGGAGGTAGTGACAA | CCTCCAATGGATCCTCGTTA | 135 |
表1 RT-qPCR所用的引物
Table 1 Primers for RT-qPCR
| 基因 Gene | 正向引物 Forward primer (5'-3') | 反向引物 Reverse primer (5'-3') | 长度Length (bp) |
|---|---|---|---|
| CtPMA1 | TTGACCAATCTGCCCTCA | CCTTCTGGAAATGCCCAA | 189 |
| CtPMA2 | CCACCAGATTGGCAAGACT | GGCATCAGCAGGAACGAT | 231 |
| CtPMA3 | AGATGCCGCCGTGTTAGT | GTCAAAGCCGACTGGTCAA | 117 |
| CtPMA4 | AGGGATGGGAGATGGTCTG | GCTTCTTGGTGACTGGTAAGG | 169 |
| CtPMA5 | TGACGACTGATGATGGTAACC | CAATCTGGAGGCTTTCCC | 181 |
| CtPMA6 | GCAGCCATTCTCGTTCCA | AGGGTGCTTTGTCACAGGA | 144 |
| CtPMA7 | GGAATCCTCTCTCGTGGGT | GCAAGCATCTTCTTCGCTC | 242 |
| CtPMA8 | GGTGCTTCTTCTCATAAACTCG | ACGATGTCTCCTGGAACCA | 162 |
| CtPMA9 | TGAAGGCATTGATAACCTGC | ACACCAGCGGCAAATACC | 246 |
| CtPMA10 | GGCGAGTCTATTCCAGTAACC | GATTGGTGTTCTCCACAAGG | 148 |
| 18S | ACACGGGGAGGTAGTGACAA | CCTCCAATGGATCCTCGTTA | 135 |

图2 CtPMA基因家族蛋白序列比对分析蓝色代表同源性≥50%,玫红色代表同源性≥75%,深蓝色代表同源性=100%;红色方框代表Cation ATPase N结构域,绿色方框代表E1_E2 ATPase结构域,亮蓝色方框代表HAD-superfamily hydrolase, subfamily ⅢA结构域
Fig. 2 Alignment analysis of protein sequencesin CtPMA gene familyBlue indicates ≥50% homology, rose red indicates ≥75% homology, and dark blue indicates homology = 100%; red boxes indicate cation ATPase N structural domains, green boxes indicate E1_E2 ATPase structural domains, and bright blue boxes indicate HAD-superfamily hydrolase, subfamily IIIA structural domains
基因名称 Gene name | 基因号 Gene ID | 蛋白质长度 Protein length (aa) | 相对分子质量 Molecular mass (kD) | 等电点 Theoretical point | 亲水性系数 Hydrophilicity coefficient | 不稳定系数 Instability index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| CtPMA1 | CtAH01T0012700.1 | 886 | 97.78 | 5.75 | 0.034 | 37.94 | 细胞膜 Plasma membrane |
| CtPMA2 | CtAH03T0170800.1 | 937 | 102.76 | 6.63 | 0.054 | 39.45 | 细胞膜 Plasma membrane |
| CtPMA3 | CtAH05T0036500.1 | 858 | 93.95 | 5.26 | 0.205 | 39.82 | 细胞膜 Plasma membrane |
| CtPMA4 | CtAH05T0146800.1 | 956 | 105.47 | 6.49 | 0.069 | 33.32 | 细胞膜 Plasma membrane |
| CtPMA5 | CtAH05T0252100.1 | 785 | 85.23 | 5.34 | 0.150 | 36.06 | 细胞膜 Plasma membrane |
| CtPMA6 | CtAH06T0267900.1 | 906 | 99.49 | 6.20 | 0.034 | 33.52 | 细胞膜 Plasma membrane |
| CtPMA7 | CtAH09T0032200.1 | 911 | 100.36 | 5.98 | 0.122 | 35.71 | 细胞膜 Plasma membrane |
| CtPMA8 | CtAH09T0161100.1 | 957 | 105.49 | 6.03 | 0.116 | 38.33 | 细胞膜 Plasma membrane |
| CtPMA9 | CtAH11T0113500.1 | 889 | 97.10 | 5.87 | 0.068 | 36.90 | 细胞膜 Plasma membrane |
| CtPMA10 | CtAH12T0151100.1 | 958 | 105.52 | 7.91 | 0.087 | 36.00 | 细胞膜 Plasma membrane |
表2 CtPMA基因家族成员理化性质
Table 2 Physicochemical properties of CtPMA gene family
基因名称 Gene name | 基因号 Gene ID | 蛋白质长度 Protein length (aa) | 相对分子质量 Molecular mass (kD) | 等电点 Theoretical point | 亲水性系数 Hydrophilicity coefficient | 不稳定系数 Instability index | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|---|
| CtPMA1 | CtAH01T0012700.1 | 886 | 97.78 | 5.75 | 0.034 | 37.94 | 细胞膜 Plasma membrane |
| CtPMA2 | CtAH03T0170800.1 | 937 | 102.76 | 6.63 | 0.054 | 39.45 | 细胞膜 Plasma membrane |
| CtPMA3 | CtAH05T0036500.1 | 858 | 93.95 | 5.26 | 0.205 | 39.82 | 细胞膜 Plasma membrane |
| CtPMA4 | CtAH05T0146800.1 | 956 | 105.47 | 6.49 | 0.069 | 33.32 | 细胞膜 Plasma membrane |
| CtPMA5 | CtAH05T0252100.1 | 785 | 85.23 | 5.34 | 0.150 | 36.06 | 细胞膜 Plasma membrane |
| CtPMA6 | CtAH06T0267900.1 | 906 | 99.49 | 6.20 | 0.034 | 33.52 | 细胞膜 Plasma membrane |
| CtPMA7 | CtAH09T0032200.1 | 911 | 100.36 | 5.98 | 0.122 | 35.71 | 细胞膜 Plasma membrane |
| CtPMA8 | CtAH09T0161100.1 | 957 | 105.49 | 6.03 | 0.116 | 38.33 | 细胞膜 Plasma membrane |
| CtPMA9 | CtAH11T0113500.1 | 889 | 97.10 | 5.87 | 0.068 | 36.90 | 细胞膜 Plasma membrane |
| CtPMA10 | CtAH12T0151100.1 | 958 | 105.52 | 7.91 | 0.087 | 36.00 | 细胞膜 Plasma membrane |
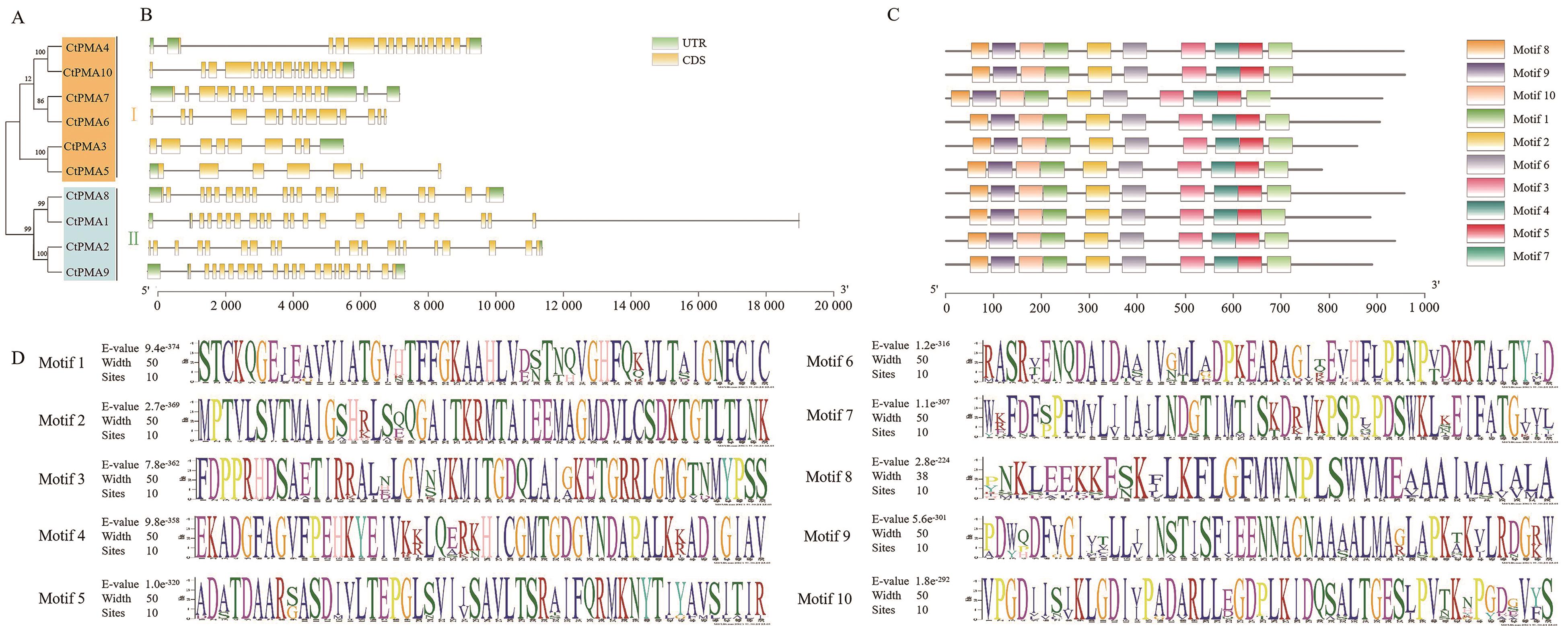
图4 CtPMAs基因结构和保守基序分析A:CtPMA基因家族的系统进化树;B:CtPMAs基因结构,其中外显子、内含子和5'/3'非翻译区分别用黄色方框、黑色线条和绿色方框标记;C和D:CtPMA基因家族的保守基序分布
Fig. 4 Gene structure and conserved motif of CtPMAsA: Phylogenetic tree of CtPMA gene family in safflower. B: Gene structure of CtPMA gene family in safflower, the yellow box refer to exons, the black lines to introns, and green box to 5'/3' untranslated regions. C and D: Distribution of conserved motifs of CtPMA gene family

图5 CtPMAs基因启动子顺式作用元件分析A:CtPMAs起始密码子上游2 000 bp启动子序列中顺式作用元件鉴定;B:各种顺式作用元件的分析
Fig. 5 Analysis of cis-acting elements in the promoter of CtPMAsA: Identification of cis-acting elements in the promoter sequence 2 000 bp upstream of the start codon of CtPMAs. B: Analysis of various cis-acting elements
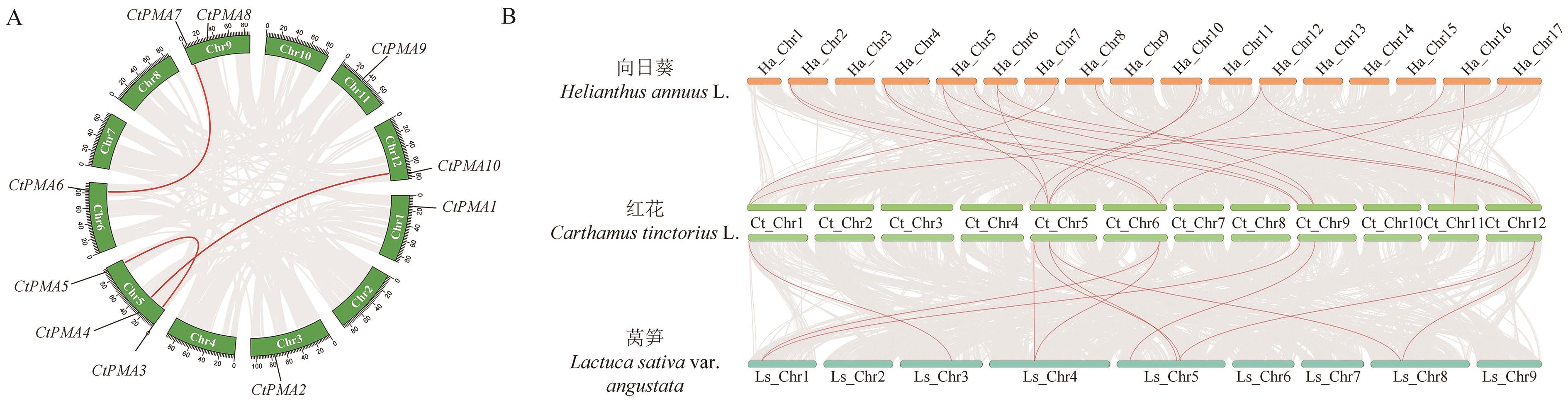
图6 CtPMAs共线性分析A:红花PMA基因家族共线性分析;B:PMA基因家族在不同物种间的比较共线性图
Fig. 6 Collinearity analysis of CtPMAsA: Collinearity analysis of PMAs gene family in safflower. B: Comparative collinearity analysis of PMA gene familyin different species
基因对 Gene pair | 复制类型 Type of duplication | Ka | Ks | Ka/Ks | 选择压力类型 Type of selective pressure |
|---|---|---|---|---|---|
| CtPMA3/CtPMA5 | 片段重复Segmental duplication | 0.130 145 43 | 1.213 732 375 | 0.107 227 452 | 纯化选择Purifying selection |
| CtPMA4/CtPMA10 | 片段重复Segmental duplication | 0.029 828 973 | 0.740 740 112 | 0.040 269 148 | 纯化选择Purifying selection |
| CtPMA6/CtPMA7 | 片段重复Segmental duplication | 0.062 455 14 | 1.531 402 62 | 0.040 782 965 | 纯化选择Purifying selection |
表4 CtPMA基因物种内共线性关系
Table 4 Interspecies collinearity relationship among CtPMA genes
基因对 Gene pair | 复制类型 Type of duplication | Ka | Ks | Ka/Ks | 选择压力类型 Type of selective pressure |
|---|---|---|---|---|---|
| CtPMA3/CtPMA5 | 片段重复Segmental duplication | 0.130 145 43 | 1.213 732 375 | 0.107 227 452 | 纯化选择Purifying selection |
| CtPMA4/CtPMA10 | 片段重复Segmental duplication | 0.029 828 973 | 0.740 740 112 | 0.040 269 148 | 纯化选择Purifying selection |
| CtPMA6/CtPMA7 | 片段重复Segmental duplication | 0.062 455 14 | 1.531 402 62 | 0.040 782 965 | 纯化选择Purifying selection |
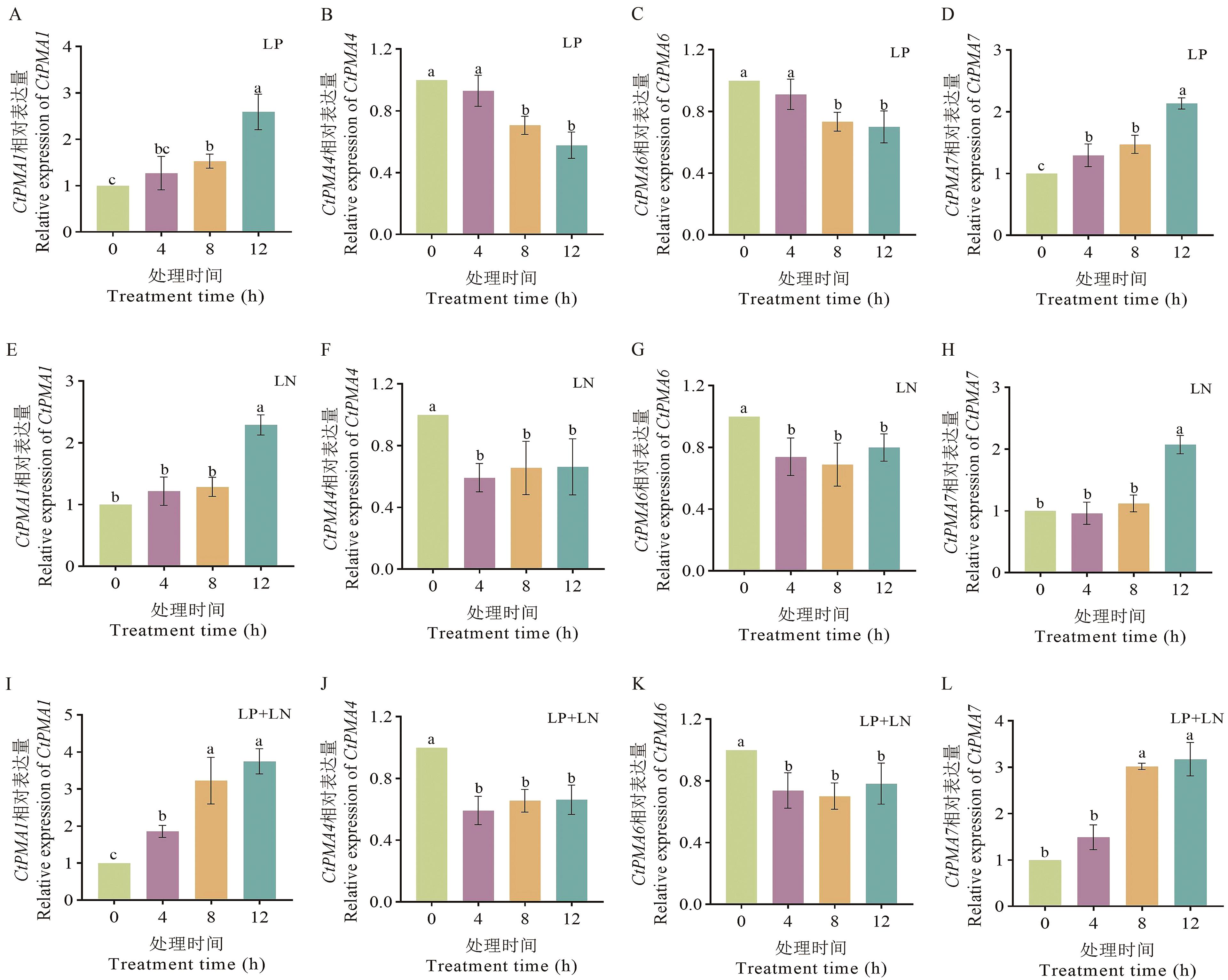
图8 CtPMA基因在低氮和磷胁迫下的响应LP:低磷胁迫,LN:低氮胁迫,LP+LN:同时低氮、磷胁迫。数值为平均值±标准差(n=6),利用Duncan’s做方差分析,不同字母表示各组之间差异显著(P<0.05)
Fig. 8 Responses of CtPMA genes under low phosphorus and low nitrogen stressLP, low phosphorus stress; LN, low nitrogen stress; LP+LN, low nitrogen and phosphorus stress. The data indicate mean ± standard deviation (n=6); different letters indicate significant differences between groups (P<0.05)
| [28] | 刘健健, 刘俊丽, 季敏杰, 等. 番茄质膜H+-ATPase家族基因的鉴定和表达分析 [J]. 浙江农林大学学报, 2016, 33(5): 734-741. |
| Liu JJ, Liu JL, Ji MJ, et al. Identification and expression analysis of tomato plasma membrane H+-ATPase family genes [J]. Journal of Zhejiang A&F University, 2016, 33(5): 734-741. | |
| [29] | 顾家家. 蓝莓质膜H+-ATPases基因家族响应非酸性根际胁迫的生物学功能研究 [D]. 金华:浙江师范大学, 2022. |
| Gu JJ. Functional analysis of plasma membrane H+-ATPase gene family in response to non-acidic stress in blueberry [D]. Jinhua: Zhejiang Normal University, 2022. | |
| [30] | 王天禧, 杨炳松, 潘荣君, 等. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究 [J]. 生物技术通报, 2025, 41(4): 176-187. |
| Wang TX, Yang BS, Pan RJ, et al. Identification of the apple PLATZ gene family and functional study of the MdPLATZ9 Gene [J]. Biotechnol Bull, 2025, 41(4): 176-187. | |
| [31] | Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and Cis-acting regulatory elements [J]. Plant Sci, 2014, 217/218: 109-119. |
| [32] | Schaller A, Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants [J]. Plant Cell, 1999, 11(2): 263-272. |
| [33] | Liu JL, Liu JJ, Chen AQ, et al. Analysis of tomato plasma membrane H(+)-ATPase gene family suggests a mycorrhiza-mediated regulatory mechanism conserved in diverse plant species [J]. Mycorrhiza, 2016, 26(7): 645-656. |
| [34] | Haruta M, Sussman MR. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis [J]. Plant Physiol, 2012, 158(3): 1158-1171. |
| [35] | Hoffmann RD, Olsen LI, Ezike CV, et al. Roles of plasma membrane proton ATPases AHA2 and AHA7 in normal growth of roots and root hairs in Arabidopsis thaliana [J]. Physiol Plant, 2019, 166(3): 848-861. |
| [36] | Vitart V, Baxter I, Doerner P, et al. Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis [J]. Plant J, 2001, 27(3): 191-201. |
| [37] | Santi S, Locci G, Monte R, et al. Induction of nitrate uptake in maize roots: expression of a putative high-affinity nitrate transporter and plasma membrane H+-ATPase isoforms [J]. J Exp Bot, 2003, 54(389): 1851-1864. |
| [1] | Zhang LL, Tian K, Tang ZH, et al. Phytochemistry and pharmacology of Carthamus tinctorius L [J]. Am J Chin Med, 2016, 44(2): 197-226. |
| [2] | 任超翔, 吴沂芸, 唐小慧, 等. 红花的起源与产地变迁 [J]. 中国中药杂志, 2017, 42(11): 2219-2222. |
| Ren CX, Wu YY, Tang XH, et al. Safflower's origin and changes of producing areas [J]. China J Chin Mater Med, 2017, 42(11): 2219-2222. | |
| [3] | Delshad E, Yousefi M, Sasannezhad P, et al. Medical uses of Carthamus tinctorius L. (safflower): a comprehensive review from traditional medicine to modern medicine [J]. Electron Physician, 2018, 10(4): 6672-6681. |
| [4] | 王佐梅, 肖洪彬, 李雪莹, 等. 中药红花的药理作用及临床应用研究进展 [J]. 中华中医药杂志, 2021, 36(11): 6608-6611. |
| Wang ZM, Xiao HB, Li XY, et al. Research progress on pharmacological actions and clinical applications of Carthami Flos [J]. China J Tradit Chin Med Pharm, 2021, 36(11): 6608-6611. | |
| [5] | 王兆木, 李文生, 吴庆红, 等. 氮磷不同用量及配比对红花产量的研究 [J]. 新疆农业科学, 2003, 40(3): 148-150. |
| Wang ZM, Li WS, Wu QH, et al. Amount of nitrogen and phosphorous fertilizer and its mixed ratio [J]. Xinjiang Agric Sci, 2003, 40(3): 148-150. | |
| [6] | 胡喜巧, 陈红芝, 邬佩宏, 等. 氮磷钾配施对红花产量及其有效成分在器官中分配的影响 [J]. 干旱地区农业研究, 2023, 41(1): 53-62. |
| Hu XQ, Chen HZ, Wu PH, et al. Effects of combined application of nitrogen, phosphorus and potassium fertilizers on yield and distribution of active ingredient in organs of C. tinctorius [J]. Agricultural Research in the Arid Areas, 2023, 41(1): 53-62. | |
| [7] | Li Y, Zeng H, Xu F, Yan F, Xu W. H+-ATPases in plant growth and stress responses[J]. Annu Rev Plant Biol, 2022, 73: 495-521. |
| [8] | Ding M, Zhang MX, Zeng HQ, et al. Molecular basis of plasma membrane H+-ATPase function and potential application in the agricultural production [J]. Plant Physiol Biochem, 2021, 168: 10-16. |
| [9] | 赵敏华, 杨王敏, 梁毅, 等. 植物质膜H+-ATPase研究进展 [J/OL]. 分子植物育种, 2022: 1-17. . |
| Zhao MH, Yang WM, Liang Y, et al. Research Progress of Plant Plasma Membrane H+-ATPase [J/OL]. Molecular Plant Breeding, 2022: 1-17. . | |
| [10] | 周思婕, 张敏, 王平. 植物质膜H+-ATP酶对环境胁迫因子的响应研究进展 [J]. 应用与环境生物学报, 2021, 27(2): 485-494. |
| Zhou SJ, Zhang M, Wang P. Response of plant plasma membrane H+-ATPase to environmental stress factors: a review [J]. Chin J Appl Environ Biol, 2021, 27(2): 485-494. | |
| [11] | Li J, Guo Y, Yang YQ. The molecular mechanism of plasma membrane H+-ATPases in plant responses to abiotic stress [J]. J Genet Genomics, 2022, 49(8): 715-725. |
| [12] | 韩秀丽. 拟南芥内源有机小分子物质调控质膜H+-ATPase活性的研究 [D]. 北京: 中国农业大学, 2020. |
| Han XL. Regulation of plasma membrane H+-ATPase activity by endogenous metabolites in Arabidopsis thaliana [D]. Beijing: China Agricultural University, 2020. | |
| [13] | Minami A, Takahashi K, Inoue SI, et al. Brassinosteroid induces phosphorylation of the plasma membrane H+-ATPase during hypocotyl elongation in Arabidopsis thaliana [J]. Plant Cell Physiol, 2019, 60(5): 935-944. |
| [14] | Pertl-Obermeyer H, Gimeno A, Kuchler V, et al. pH modulates interaction of 14-3-3 proteins with pollen plasma membrane H+-ATPases independently from phosphorylation [J]. J Exp Bot, 2022, 73(1): 168-181. |
| [15] | Quaggiotti S, Ruperti B, Borsa P, et al. Expression of a putative high-affinity NO3 - transporter and of an H+-ATPase in relation to whole plant nitrate transport physiology in two maize genotypes differently responsive to low nitrogen availability [J]. J Exp Bot, 2003, 54(384): 1023-1031. |
| [16] | Shen H, Chen JH, Wang ZY, et al. Root plasma membrane H+-ATPase is involved in the adaptation of soybean to phosphorus starvation [J]. J Exp Bot, 2006, 57(6): 1353-1362. |
| [17] | Baxter I, Tchieu J, Sussman MR, et al. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice [J]. Plant Physiol, 2003, 132(2): 618-628. |
| [38] | Sakuraba Y, Kanno S, Mabuchi A, et al. A phytochrome-B-mediated regulatory mechanism of phosphorus acquisition [J]. Nat Plants, 2018, 4(12): 1089-1101. |
| [39] | Zhang RP, Liu G, Wu N, et al. Adaptation of plasma membrane H+-ATPase and H+ pump to P deficiency in rice roots [J]. Plant Soil, 2011, 349(1): 3-11. |
| [40] | Yan F, Zhu YY, Müller C, et al. Adaptation of H+-pumping and plasma membrane H+-ATPase activity in proteoid roots of white lupin under phosphate deficiency [J]. Plant Physiol, 2002, 129(1): 50-63. |
| [41] | Feng HM, Fan XR, Miller AJ, et al. Plant nitrogen uptake and assimilation: regulation of cellular pH homeostasis [J]. J Exp Bot, 2020, 71(15): 4380-4392. |
| [42] | Sperandio MVL, Santos LA, Bucher CA, et al. Isoforms of plasma membrane H(+)-ATPase in rice root and shoot are differentially induced by starvation and resupply of NO₃⁻ or NH₄+ [J]. Plant Sci, 2011, 180(2): 251-258. |
| [43] | Loss Sperandio MV, Santos LA, Huertas Tavares OC, et al. Silencing the Oryza sativa plasma membrane H+-ATPase isoform OsA2 affects grain yield and shoot growth and decreases nitrogen concentration [J]. J Plant Physiol, 2020, 251: 153220. |
| [44] | Ding M, Zhang MX, Wang ZH, et al. Overexpression of a plasma membrane H+-ATPase gene OSA1 stimulates the uptake of primary macronutrients in rice roots [J]. Int J Mol Sci, 2022, 23(22): 13904. |
| [45] | Zhang MX, Wang Y, Chen X, et al. Plasma membrane H+-ATPase overexpression increases rice yield via simultaneous enhancement of nutrient uptake and photosynthesis [J]. Nat Commun, 2021, 12(1): 735. |
| [18] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a "one for all, all for one" bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [19] | Hong YQ, Lv YX, Zhang JY, et al. The safflower MBW complex regulates HYSA accumulation through degradation by the E3 ligase CtBB1 [J]. J Integr Plant Biol, 2023, 65(5): 1277-1296. |
| [20] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method [J]. Methods, 2001, 25(4): 402-408. |
| [21] | Tian W, Huang X, Ouyang XH. Genome-wide prediction of activating regulatory elements in rice by combining STARR-seq with FACS [J]. Plant Biotechnol J, 2022, 20(12): 2284-2297. |
| [22] | Roth C, Liberles DA. A systematic search for positive selection in higher plants (Embryophytes) [J]. BMC Plant Biol, 2006, 6: 12. |
| [23] | 闫文惠, 陆婷婷, 王焕然, 等. 质膜H+-ATPase影响植物生长发育及逆境响应研究进展 [J]. 植物生理学报, 2024, 60(10): 1487-1501. |
| Yan WH, Lu TT, Wang HR, et al. Research progress of plasma membrane H+-ATPase functioned in plant growth and development and its response to abiotic stress [J]. Plant Physiol Journal, 2024, 60(10): 1487-1501. | |
| [24] | Zeng HQ, Chen HY, Zhang MX, et al. Plasma membrane H+-ATPases in mineral nutrition and crop improvement [J]. Trends Plant Sci, 2024, 29(9): 978-994. |
| [25] | Arango M, Gévaudant F, Oufattole M, et al. The plasma membrane proton pump ATPase: the significance of gene subfamilies [J]. Planta, 2003, 216(3): 355-365. |
| [26] | Huang Y, Cao HS, Yang L, et al. Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae [J]. J Exp Bot, 2019, 70(20): 5879-5893. |
| [27] | Oufattole M, Arango M, Boutry M. Identification and expression of three new Nicotiana plumbaginifolia genes which encode isoforms of a plasma-membrane H(+)-ATPase, and one of which is induced by mechanical stress [J]. Planta, 2000, 210(5): 715-722. |
| [1] | 赖诗雨, 梁巧兰, 魏列新, 牛二波, 陈应娥, 周鑫, 杨思正, 王博. NbJAZ3在苜蓿花叶病毒侵染本氏烟过程中的作用[J]. 生物技术通报, 2025, 41(8): 186-196. |
| [2] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| [3] | 李志强, 罗正乾, 徐琳黎, 周国慧, 屈丝雨, 刘恩良, 顼东婷. 基于T2T基因组鉴定大豆R2R3-MYB基因家族及干旱和盐胁迫下的表达分析[J]. 生物技术通报, 2025, 41(5): 141-152. |
| [4] | 王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175. |
| [5] | 班秋艳, 赵鑫月, 迟文静, 黎俊生, 王琼, 夏瑶, 梁丽云, 贺巍, 李叶云, 赵广山. 茶树光敏色素互作因子CsPIF3a的克隆及其与光温逆境的响应[J]. 生物技术通报, 2025, 41(4): 256-265. |
| [6] | 卢勇杰, 夏海乾, 李永铃, 张文建, 余婧, 赵会纳, 王兵, 许本波, 雷波. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析[J]. 生物技术通报, 2025, 41(4): 266-277. |
| [7] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| [8] | 王琛, 刘国梅, 陈畅, 张晋龙, 姚琳, 孙璇, 杜春芳. 白菜型油菜CCDs家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(3): 161-170. |
| [9] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| [10] | 李明, 刘祥宇, 王益娜, 和四梅, 沙本才. 紫金龙异紫堇定生物合成相关6-OMT基因克隆与功能表征[J]. 生物技术通报, 2025, 41(2): 309-320. |
| [11] | 王子傲, 田瑞, 崔永梅, 白羿雄, 姚晓华, 安立昆, 吴昆仑. 青稞HvnJAZ4的生物信息学和表达模式分析[J]. 生物技术通报, 2025, 41(1): 173-185. |
| [12] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [13] | 吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91. |
| [14] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [15] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||