








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 106-114.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0780
• 研究报告 • 上一篇
陈晓军1( ), 惠建1, 马洪文2, 白海波1, 钟楠1, 李稼润1, 樊云芳3
), 惠建1, 马洪文2, 白海波1, 钟楠1, 李稼润1, 樊云芳3
收稿日期:2024-08-14
出版日期:2025-04-26
发布日期:2025-04-25
作者简介:陈晓军,博士,副研究员,研究方向 :植物分子生物学;E-mail: smallgene@126.com;陈晓军同为本文
基金资助:
CHEN Xiao-jun1( ), HUI Jian1, MA Hong-wen2, BAI Hai-Bo1, ZHONG Nan1, LI Jia-run1, FAN Yun-fang3
), HUI Jian1, MA Hong-wen2, BAI Hai-Bo1, ZHONG Nan1, LI Jia-run1, FAN Yun-fang3
Received:2024-08-14
Published:2025-04-26
Online:2025-04-25
摘要:
目的 以宁夏耐盐品种宁粳56为实验材料,定点编辑乙酰乳酸合酶ALS基因,创制具有抗除草剂特性的耐盐水稻新种质资源。 方法 以ALS基因171和190两个位点为靶位点,利用胞嘧啶碱基编辑器BE3,构建单碱基编辑载体OsALS171-SpG-eBE3和OsALS190-SpG-eBE3,通过农杆菌介导进行受体材料的基因转化获得基因编辑植株,采用靶位点测序的方法进行基因型鉴定;建立了除草剂耐受系统并对其除草剂的耐受程度进行了快速鉴定。 结果 经T1代靶位点测序验证,获得ALSP171F 2个突变纯合株系;ALSE187K R190H、ALSE187K、ALSV188I、ALSR190H、ALSD201N 5种突变类型纯合株系各1株。相对野生型受体材料,ALSP171F 2个突变纯合株系对双草醚有较强的耐受力,而ALSE187K R190H、ALSE187K、ALSV188I、ALSR190H、ALSD201N 5种突变纯合植株虽然对双草醚有一定的耐受,但耐受程度不显著。 结论 利用SpG胞嘧啶碱基编辑器BE3获得具有抗草剂特性的基因编辑材料,且能够稳定遗传,ALSP171F位点突变对双草醚除草剂表现较强抗性,是培育抗除草剂水稻的优先位点。
陈晓军, 惠建, 马洪文, 白海波, 钟楠, 李稼润, 樊云芳. 利用单碱基基因编辑技术创制OsALS抗除草剂水稻种质资源[J]. 生物技术通报, 2025, 41(4): 106-114.
CHEN Xiao-jun, HUI Jian, MA Hong-wen, BAI Hai-Bo, ZHONG Nan, LI Jia-run, FAN Yun-fang. Creating Rice Gerplasm Resources OsALS Rsistant to Herbicide through Single Base Gene Editing Technology[J]. Biotechnology Bulletin, 2025, 41(4): 106-114.
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途 Purpose |
|---|---|---|
| OsALS171-gRNA-F | ggcaGTCCCCCGCCGCATGAT | OsALS171位点基因编辑载体构建 |
| OsALS171-gRNA-R | aaacATCATGCGGCGGGGGAC | |
| OsALS190-gRNA-F | ggcaGGAGCGGGTGACCTCGACTA | OsALS190位点基因编辑载体构建 |
| OsALS190-gRNA-R | aaacTAGTCGAGGTCACCCGCTCC | |
| OsALS-dect-F | GCTCGGCGGTGTCCCCGGTC | 两个靶位点检测 |
| OsALS-dect-R | CCGGCACGGCCATCTGCTGC |
表1 引物设计列表
Table 1 List of designed primers
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途 Purpose |
|---|---|---|
| OsALS171-gRNA-F | ggcaGTCCCCCGCCGCATGAT | OsALS171位点基因编辑载体构建 |
| OsALS171-gRNA-R | aaacATCATGCGGCGGGGGAC | |
| OsALS190-gRNA-F | ggcaGGAGCGGGTGACCTCGACTA | OsALS190位点基因编辑载体构建 |
| OsALS190-gRNA-R | aaacTAGTCGAGGTCACCCGCTCC | |
| OsALS-dect-F | GCTCGGCGGTGTCCCCGGTC | 两个靶位点检测 |
| OsALS-dect-R | CCGGCACGGCCATCTGCTGC |

图3 植物单碱基基因编辑载体靶位点测序A:OsALS171-SpG-eBE3载体靶位点测序;B:OsALS190-SpG-eBE3载体靶位点测序
Fig. 3 Sequencing of gRNA in plant single base editing vectorA: Sequencing of target sites in vector OsALS171-SpG-eBE3. B: Sequencing of target sites in vector OsALS190-SpG-eBE3
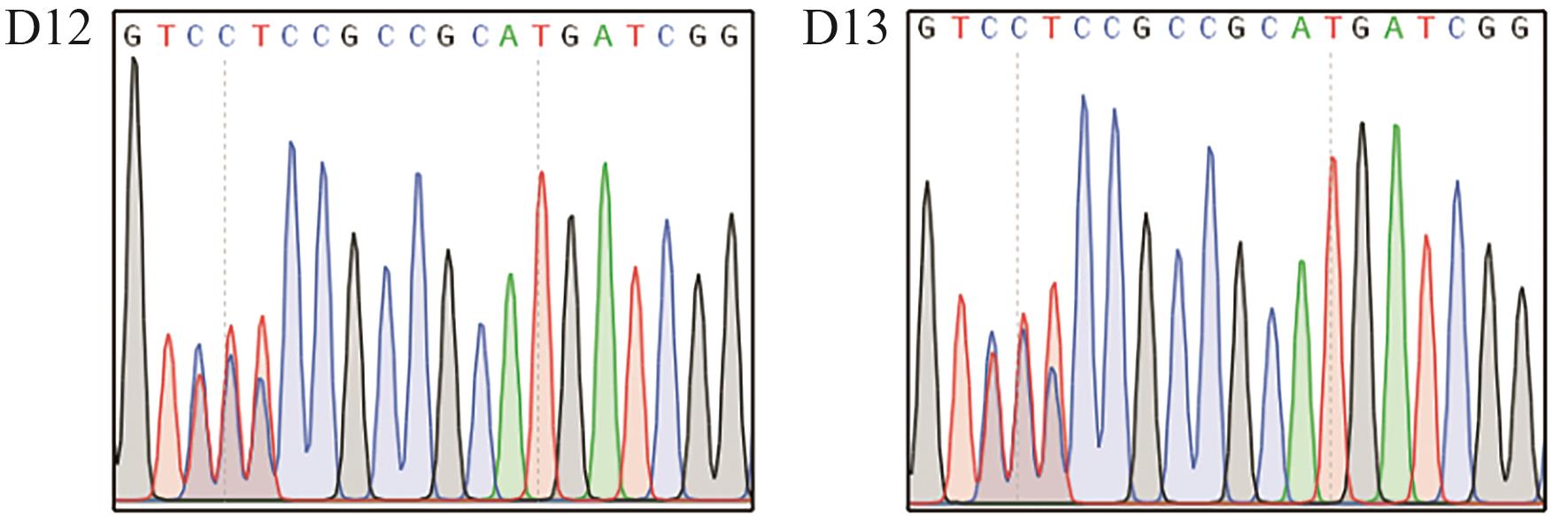
图4 OsALS171-SpG-eBE3 T0代编辑结果图中胞嘧啶碱基C和胸腺嘧啶碱基T的套峰,说明该位点进行了定点编辑
Fig. 4 Editing results of OsALS171-SpG-eBE3 in T0 plantsThe overlapping peak of cytosine base C and thymine base T indicates edited bases in target sites
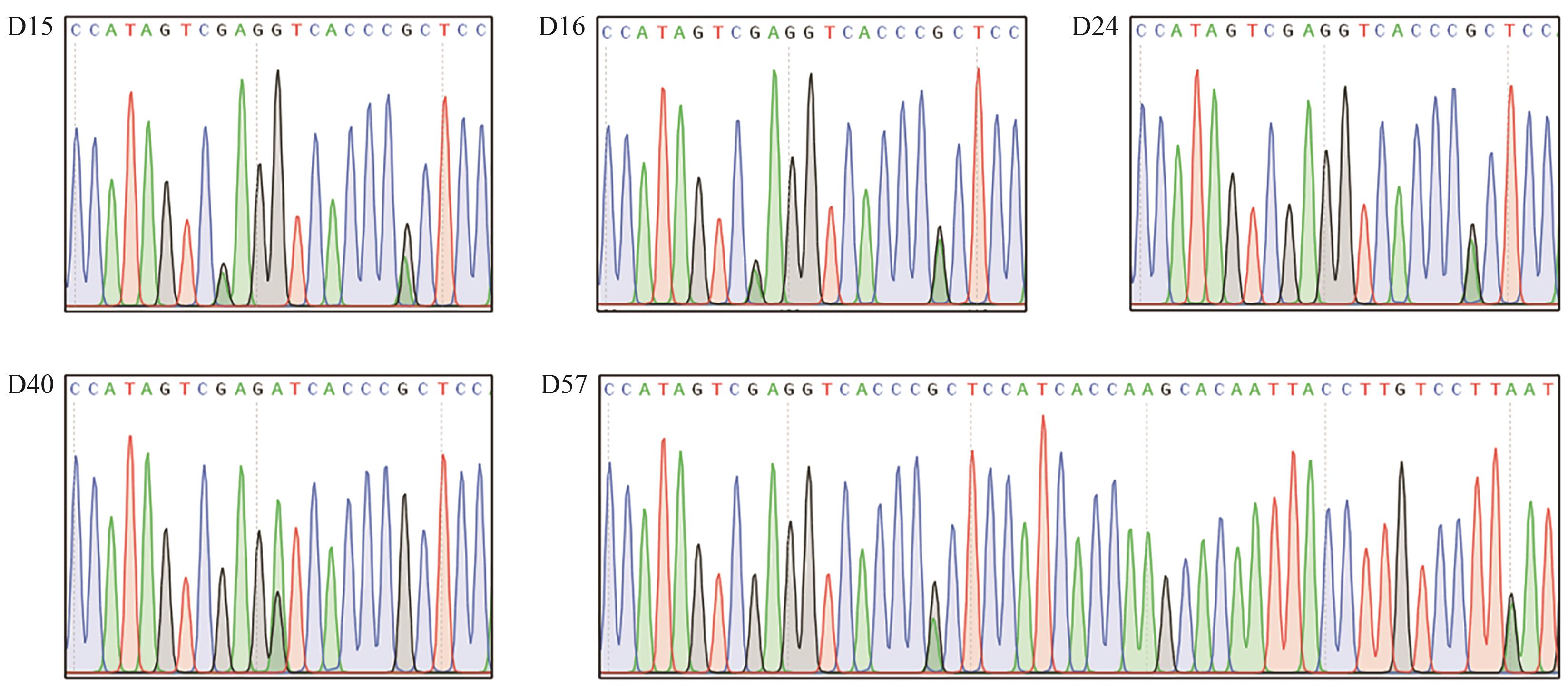
图5 OsALS190-SpG-eBE3 T0代编辑结果图中出现套峰的位置均表明进行了定点编辑
Fig. 5 Editing results of OsALS190-SpG-eBE3 in T0 plantsThe overlapping peak indicates edited bases in target sites

图6 OsALS171-SpG-eBE3 T1代编辑结果D12-9,D13-4植株OsALS基因171位氨基酸由P突变为F
Fig. 6 Editing results of OsALS171-SpG-eBE3 in T1 plantsP amino acids mutated F amino acids at 171 of OsALS in D12-9 and D13-4 plants
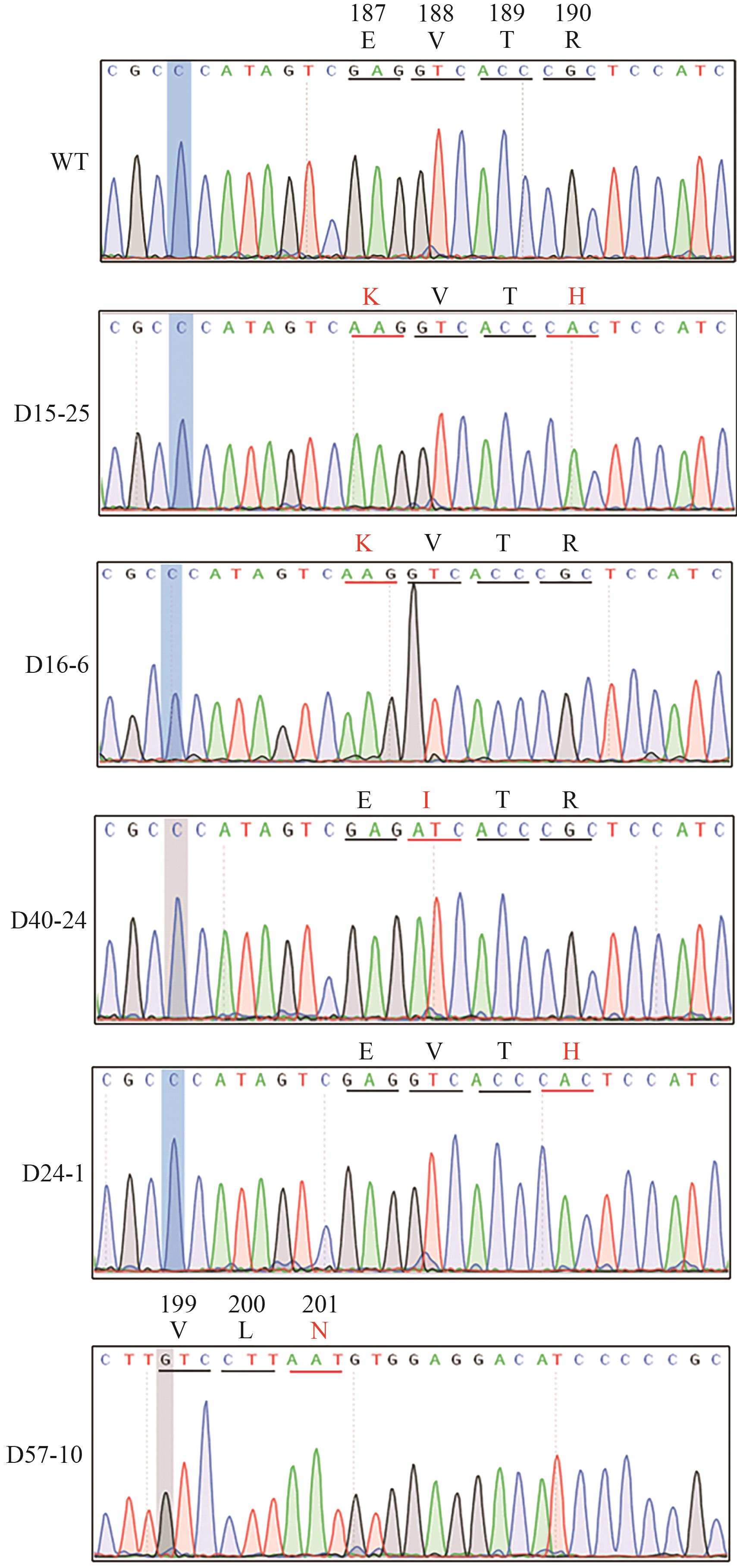
图7 OsALS190-SpG-eBE3 T1代编辑结果D15-25植株OsALS基因187位氨基酸由E突变为K,190位氨基酸由R突变为H;D16-6植株OsALS基因187位氨基酸由E突变为K;D40-24植株OsALS基因188位氨基酸由V突变为I;D24-1植株OsALS基因190位氨基酸由R突变为H;D57-10植株OsALS基因201位氨基酸由D突变为N
Fig. 7 Editing results of OsALS171-SpG-eBE3 in T1 plantsE amino acids mutated K amino acids at 187 of OsALS and R amino acids mutated H amino acids at 190 of OsALS in D15-25 plants. E amino acids mutated K amino acids at 187 of OsALS in D16-6 plants. V amino acids mutated I amino acids at 188 of OsALS in D40-24 plants. R amino acids mutated H amino acids at 190 of OsALS in D24-1 plants. D amino acids mutated N amino acids at 201 of OsALS in D57-10 plants

图9 基因编辑纯合体对双草醚的抗性鉴定A:野生型受体;B:D12-9纯合植株;C:D13-4纯合植株;D:D15-25纯合植株;E:D16-6纯合植株;F:D40-24纯合植株;G:D24-1纯合植株;H:D57-10纯合植株。每组中,左到右双草醚浓度分别为0,0.2,0.4 μmol/L
Fig. 9 Characterization of edited homozygote for resistance to bispyribac-sodiumA: Wild type receptor plants; B: D12-9 homozygote plants; C: D13-4 homozygote plants; D: D15-25 homozygote plants; E: D16-6 homozygote plants; F: D40-24 homozygote plants; G: D24-1 homozygote plants; H: D57-10 homozygote plants. In each group, the left-to-right bispyribac-sodium concentrations are 0, 0.2, and 0.4 μmol/L, respectively
| 1 | 曾波. 近30年来我国水稻主要品种更新换代历程浅析 [J]. 作物杂志, 2018(3): 1-7. |
| Zeng B. Renovation of main cultivated rice varieties in China in the past 30 years [J]. Crops, 2018(3): 1-7. | |
| 2 | 黄健, 朱安, 汪浩, 等. 水直播和旱直播对水稻产量与品质的影响综述 [J]. 江苏农业科学, 2020, 48(16): 67-73. |
| Huang J, Zhu A, Wang H, et al. Effects of water direct-seeding and dry direct-seeding on yield and quality of rice: a review [J]. Jiangsu Agric Sci, 2020, 48(16): 67-73. | |
| 3 | 章秀福, 朱德峰. 中国直播稻生产现状与前景展望 [J]. 中国稻米, 1996, 2(5): 1-4. |
| Zhang XF, Zhu DF. Present situation and prospect of direct seeding rice production in China [J]. China Rice, 1996, 2(5): 1-4. | |
| 4 | 邱龙, 马崇烈, 刘博林, 等. 耐除草剂转基因作物研究现状及发展前景 [J]. 中国农业科学, 2012, 45(12): 2357-2363. |
| Qiu L, Ma CL, Liu BL, et al. Current situation of research on transgenic crops with herbicide tolerance and development prospect [J]. Sci Agric Sin, 2012, 45(12): 2357-2363. | |
| 5 | Zong Y, Liu YJ, Xue CX, et al. An engineered prime editor with enhanced editing efficiency in plants [J]. Nat Biotechnol, 2022, 40(9): 1394-1402. |
| 6 | Fartyal D, Agarwal A, James D, et al. Co-expression of P173S mutant rice EPSPS and igrA genes results in higher glyphosate tolerance in transgenic rice [J]. Front Plant Sci, 2018, 9: 144. |
| 7 | Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage [J]. Nature, 2016, 533(7603): 420-424. |
| 8 | Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage [J]. Nature, 2017, 551(7681): 464-471. |
| 9 | Tong HW, Wang XC, Liu YH, et al. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase [J]. Nat Biotechnol, 2023, 41(8): 1080-1084. |
| 10 | Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C: G-to-T: a base editors with higher efficiency and product purity [J]. Sci Adv, 2017, 3(8): eaao4774. |
| 11 | Li J, Xu RF, Qin RY, et al. Genome editing mediated by SpCas9 variants with broad non-canonical PAM compatibility in plants [J]. Mol Plant, 2021, 14(2): 352-360. |
| 12 | 辛洁, 徐小博, 王磊, 等. ALS抑制剂类除草剂的抗性研究概述 [J]. 安徽农业科学, 2019, 47(4): 18-21. |
| Xin J, Xu XB, Wang L, et al. Study on the resistance of ALS inhibitor herbicides [J]. J Anhui Agric Sci, 2019, 47(4): 18-21. | |
| 13 | Wang MG, Lu YM, Botella JR, et al. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system [J]. Mol Plant, 2017, 10(7): 1007-1010. |
| 14 | Sun YW, Zhang X, Wu CY, et al. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase [J]. Mol Plant, 2016, 9(4): 628-631. |
| 15 | Shimatani Z, Kashojiya S, Takayama M, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion [J]. Nat Biotechnol, 2017, 35(5): 441-443. |
| 16 | Kuang YJ, Li SF, Ren B, et al. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms [J]. Mol Plant, 2020, 13(4): 565-572. |
| 17 | Fan TT, Cheng YH, Wu YC, et al. High performance TadA-8e derived cytosine and dual base editors with undetectable off-target effects in plants [J]. Nat Commun, 2024, 15(1): 5103. |
| 18 | Chen YY, Wang ZP, Ni HW, et al. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis [J]. Sci China Life Sci, 2017, 60(5): 520-523. |
| 19 | Li YM, Zhu JJ, Wu H, et al. Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize [J]. Crop J, 2020, 8(3): 449-456. |
| 20 | Tian SW, Jiang LJ, Cui XX, et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing [J]. Plant Cell Rep, 2018, 37(9): 1353-1356. |
| 21 | Veillet F, Perrot L, Chauvin L, et al. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor [J]. Int J Mol Sci, 2019, 20(2): 402. |
| 22 | Wu J, Chen C, Xian GY, et al. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing [J]. Plant Biotechnol J, 2020, 18(9): 1857-1859. |
| 23 | Zhang R, Liu J, Chai Z, et al. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing [J]. Nat Plants, 2019, 5(5): 480-485. |
| 24 | 陈晓军, 吴凯伸, 惠建, 等. 基于基因编辑技术创制高亚油酸水稻材料 [J]. 中国农业科学, 2021, 54(14): 2931-2940. |
| Chen XJ, Wu KS, Hui J, et al. Production of high linoleic acid rice by genome editing [J]. Sci Agric Sin, 2021, 54(14): 2931-2940. | |
| 25 | 陈晓军, 王敬东, 宋海丽, 等. 一种简单 、极快的植物叶片DNA提取方法 [J]. 种子, 2018, 37(11): 26-29, 34. |
| Chen XJ, Wang JD, Song HL, et al. A simple and rapid method of DNA extraction from plant leaf [J]. Seed, 2018, 37(11): 26-29, 34. | |
| 26 | 余铮, 邓莉立, 谭显胜, 等. 20%双草醚可湿性粉剂对水稻直播田杂草的防除效果及安全性评价 [J]. 杂草学报, 2017, 35(3): 38-42. |
| Yu Z, Deng LL, Tan XS, et al. Efficacy and selectivity of bispyribac-sodium 20% WP in direct-seeded rice fields [J]. J Weed Sci, 2017, 35(3): 38-42. | |
| 27 | Croughan TP. Resistance to acetohydroxyacid synthase-inhibiting herbicides in rice: US20090025108 [P]. 2009-01-22. |
| 28 | Shoba D, Raveendran M, Manonmani S, et al. Development and genetic characterization of A novel herbicide (imazethapyr) tolerant mutant in rice (Oryza sativa L.) [J]. Rice, 2017, 10(1): 10. |
| 29 | Chen L, Gu G, Wang CX, et al. Trp548Met mutation of acetolactate synthase in rice confers resistance to a broad spectrum of ALS-inhibiting herbicides [J]. Crop J, 2021, 9(4): 750-758. |
| 30 | Okuzaki A, Shimizu T, Kaku K, et al. A novel mutated acetolactate synthase gene conferring specific resistance to pyrimidinyl carboxy herbicides in rice [J]. Plant Mol Biol, 2007, 64(1-2): 219-224. |
| 31 | Kawai K, Kaku K, Izawa N, et al. A novel mutant acetolactate synthase gene from rice cells, which confers resistance to ALS-inhibiting herbicides [J]. J Pestic Sci, 2007, 32(2): 89-98. |
| 32 | Zafar K, Khan MZ, Amin I, et al. Employing template-directed CRISPR-based editing of the OsALS gene to create herbicide tolerance in Basmati rice [J]. AoB Plants, 2023, 15(2): plac059. |
| 33 | Ren QR, Sretenovic S, Liu SS, et al. PAM-less plant genome editing using a CRISPR-SpRY toolbox [J]. Nat Plants, 2021, 7(1): 25-33. |
| 34 | Wang FQ, Xu Y, Li WQ, et al. Creating a novel herbicide-tolerance OsALS allele using CRISPR/Cas9-mediated gene editing [J]. Crop J, 2021, 9(2): 305-312. |
| 35 | Nishizawa-Yokoi A, Mikami M, Toki S. A universal system of CRISPR/Cas9-mediated gene targeting using all-in-one vector in plants [J]. Front Genome Ed, 2020, 2: 604289. |
| 36 | Zhang R, Chen S, Meng XB, et al. Generating broad-spectrum tolerance to ALS-inhibiting herbicides in rice by base editing [J]. Sci China Life Sci, 2021, 64(10): 1624-1633. |
| 37 | Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants [J]. Science, 2020, 368(6488): 290-296. |
| [1] | 李欣芃, 张武汉, 张莉, 舒服, 何强, 郭杨, 邓华凤, 王悦, 孙平勇. γ射线诱变创制水稻突变体及其分子鉴定[J]. 生物技术通报, 2025, 41(3): 35-43. |
| [2] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [3] | 金素奎, 国倩倩, 刘巧泉, 高继平. 一种水稻叶片基因组DNA简易提取方法[J]. 生物技术通报, 2025, 41(1): 74-84. |
| [4] | 刘文志, 贺丹, 李鹏, 傅应林, 张译心, 温华杰, 于文清. 多粘类芽胞杆菌新菌株X-11及其对番茄和水稻的促生效应[J]. 生物技术通报, 2024, 40(9): 249-259. |
| [5] | 朱诗斐, 刘敬, 张家芊, 黄文坤, 彭德良, 孔令安, 彭焕. 水稻和拟禾本科根结线虫互作分子机制研究进展[J]. 生物技术通报, 2024, 40(9): 172-180. |
| [6] | 李庆懋, 彭聪归, 齐笑含, 刘兴蕾, 李臻园, 李沁妍, 黄立钰. 促进水稻铁素吸收的野生稻内生细菌优良菌株的筛选与鉴定[J]. 生物技术通报, 2024, 40(8): 255-263. |
| [7] | 孙志勇, 杜怀东, 刘阳, 马嘉欣, 于雪然, 马伟, 姚鑫杰, 王敏, 李培富. 水稻籽粒γ-氨基丁酸含量的全基因组关联分析[J]. 生物技术通报, 2024, 40(8): 53-62. |
| [8] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| [9] | 田胜尼, 张琴, 董玉飞, 丁洲, 叶爱华, 张明珠. 酸性矿山废水对成熟期水稻根区理化因子及固氮微生物的影响[J]. 生物技术通报, 2024, 40(6): 271-280. |
| [10] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [11] | 李兴容, 谭志兵, 赵燕, 李曜魁, 赵炳然, 唐丽. 水稻低亲和性阳离子转运蛋白基因OsLCT3的克隆与功能研究[J]. 生物技术通报, 2024, 40(4): 97-109. |
| [12] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| [13] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [14] | 张超, 王子瑞, 孙亚丽, 毛馨晨, 唐家琪, 于恒秀. 水稻维生素B1合成相关基因OsTHIC的功能研究[J]. 生物技术通报, 2024, 40(2): 99-108. |
| [15] | 戚旺, 毕一凡, 轩强兵, 张新, 周慧岗, 聂圆情, 程彪彪, 张禹舜, 王俊杰, 梁卫红. 水稻OsRhoGDI1过表达影响粒型及种子活力[J]. 生物技术通报, 2024, 40(11): 152-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||