








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 47-60.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0982
收稿日期:2024-10-08
出版日期:2025-04-26
发布日期:2025-04-25
通讯作者:
费强,男,博士,教授,研究方向 :一碳合成生物制造;E-mail: feiqiang@xjtu.edu.cn作者简介:鲁天怡,女,硕士研究生,研究方向 :生物转化甲烷合成聚乳酸;E-mail: luty6099@stu.xjtu.edu.cn
基金资助:
LU Tian-yi1( ), LI Ai-peng1,2, FEI Qiang1,2(
), LI Ai-peng1,2, FEI Qiang1,2( )
)
Received:2024-10-08
Published:2025-04-26
Online:2025-04-25
摘要:
聚乳酸(polylactic acid, PLA)是以乳酸为原料聚合而成的非天然生物可降解塑料,具有优良的生物降解性,是碳达峰、碳中和背景下传统石油基塑料的重要替代品之一。作为典型的碳中和材料,PLA正逐步成为国民经济和社会发展所需的基础性大宗原材料。目前,PLA主要通过生物发酵与化学聚合相结合的工艺生产,生产过程复杂,成本较高,且存在毒性物质残留的隐患。因此,更加绿色便捷的生产方法开发成为PLA合成领域的关注热点。随着合成生物学、蛋白质工程和代谢工程的快速发展,PLA全生物合成关键酶被逐渐挖掘和改造,PLA全生物合成路径被设计和组装,以具有工业化属性的微生物为底盘构建一步合成PLA的细胞工厂成为现实,为PLA的绿色合成提供了新的解决方案。然而,生物合成法面临PLA产量低、产品性能差等问题,难以满足工业化生产的要求。因此,提高PLA生物合成效率和改善产品性能成为PLA生物合成技术开发的重点。本文首先对PLA及其合成方法进行了介绍,并系统分析了化学合成法与生物合成法各自的优势和不足;随后,总结了PLA生物合成的途径和关键酶,着重从蛋白质工程和代谢工程两个方面归纳了PLA生物合成的调控策略;最后,对PLA生物合成技术升级发展中面临的关键挑战和未来研究趋势进行了系统的分析和展望,旨在为更高效、更绿色的PLA生物合成系统的设计和开发提供有益参考。
鲁天怡, 李爱朋, 费强. 生物合成聚乳酸研究进展[J]. 生物技术通报, 2025, 41(4): 47-60.
LU Tian-yi, LI Ai-peng, FEI Qiang. Research Progress in the Biosynthesis of Polylactic Acid[J]. Biotechnology Bulletin, 2025, 41(4): 47-60.
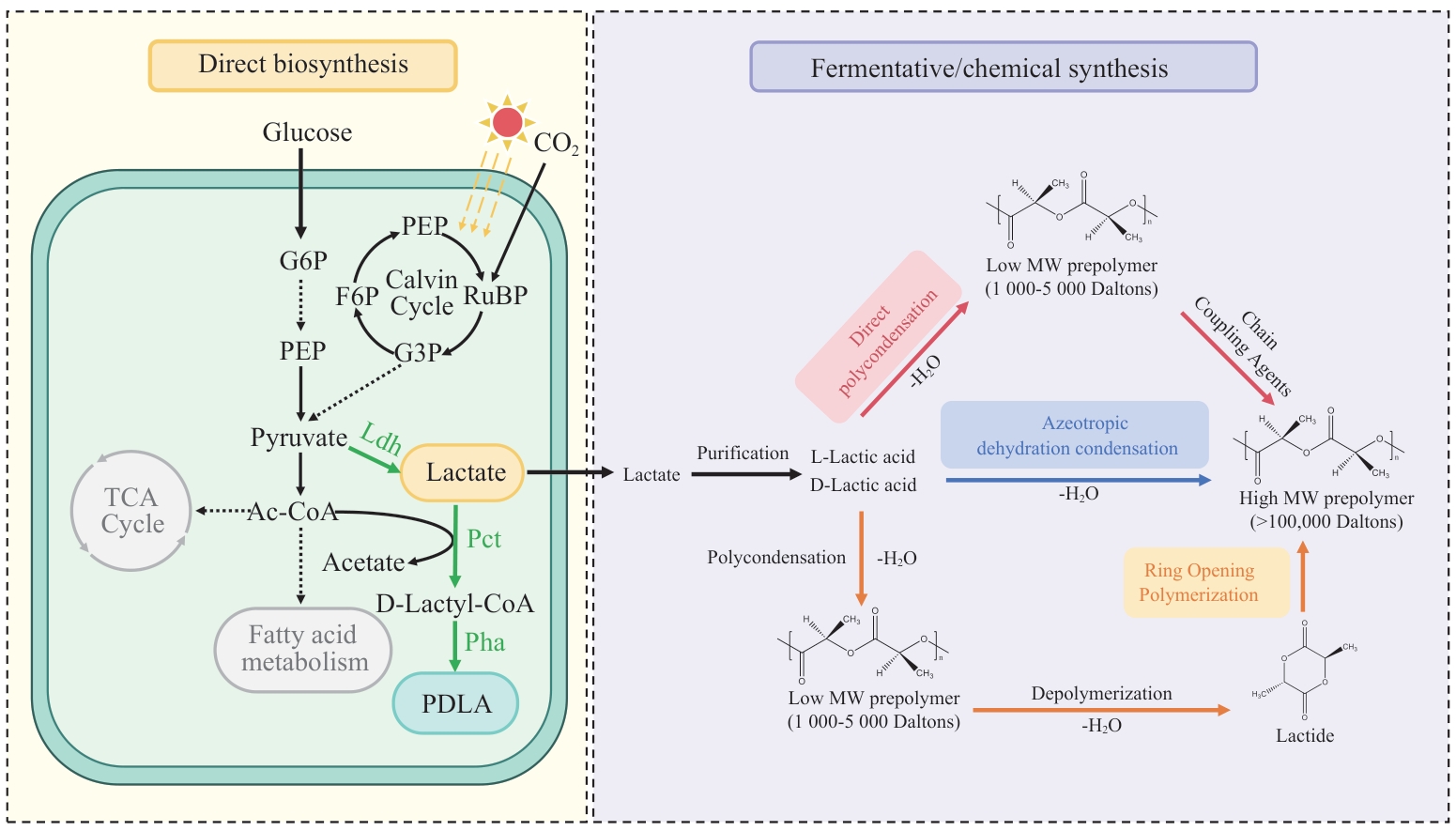
图3 PLA的直接生物合成工艺和传统生化杂合工艺的比较G6P:6-磷酸甘油醛;PEP:磷酸烯醇式丙酮酸;Ac-CoA:乙酰辅酶A;Ldh:乳酸脱氢酶;Pct:丙酰辅酶A转移酶;Pha:PHA合成酶
Fig. 3 Comparison of direct biosynthesis process and traditional biochemical hybrid process of PLAG6P: Glyceraldehyde 6-phosphate; PEP: phosphoenolpyruvate; Ac-CoA: acetyl-CoA; Ldh: lactate dehydrogenase; Pct: propionyl-CoA transferase; Pha: PHA synthase
| 方法 Method | 优点 Advantage | 缺点 Disadvantage |
|---|---|---|
直接缩合 Direct polycondensation | 1)操作简单且相对经济; 2)无需中间体的纯化,成本较低; 3)制得PLA的分子量低,降解速度快 | 1)易发生各种副反应; 2)低分子量PLA拉伸性能差,不能加工为塑料和纺织品; 3)易引入人体难以降解的杂质,不利于在医疗方面的应用[ |
共沸脱水缩合 Azeotropic dehydration condensation | 1)易除去反应中的水,使反应更易正向进行; 2)制得PLA的含水量低,分子量高 | 1)反应条件苛刻,设备和工艺复杂性较高; 2)消耗大量有机溶剂,成本高且安全性低; 3)易引入杂质和各种副反应; 4)产物提纯复杂; 5)制得的PLA易存在溶剂和催化剂残留,无法应用于医学领域[ |
开环聚合 Ring opening polymerization | 1)制得PLA的分子量高且集中; 2)制得PLA纯度高,力学性能好; 3)易调控PLA的化学结构,获得指定产物 | 1)丙交酯纯度要求高,纯化难度大; 2)合成工艺复杂、成本高、产率低 |
生物聚合 Biopolymerization | 1)反应条件温和,不易引入杂质和副反应; 2)PLA中不存在残留单体和毒性物质; 3)易生产优异对映纯度的聚合物; 4)易实现乳酸和其他单体的共聚 | 1)PLA产率低、分子量低; 2)生产成本和材料性能不可观; 3)尚处于实验室研究阶段,难以大规模应用 |
表1 PLA合成方法的分类及其优缺点比较
Table 1 Synthesis method of PLA and its advantages and disadvantages
| 方法 Method | 优点 Advantage | 缺点 Disadvantage |
|---|---|---|
直接缩合 Direct polycondensation | 1)操作简单且相对经济; 2)无需中间体的纯化,成本较低; 3)制得PLA的分子量低,降解速度快 | 1)易发生各种副反应; 2)低分子量PLA拉伸性能差,不能加工为塑料和纺织品; 3)易引入人体难以降解的杂质,不利于在医疗方面的应用[ |
共沸脱水缩合 Azeotropic dehydration condensation | 1)易除去反应中的水,使反应更易正向进行; 2)制得PLA的含水量低,分子量高 | 1)反应条件苛刻,设备和工艺复杂性较高; 2)消耗大量有机溶剂,成本高且安全性低; 3)易引入杂质和各种副反应; 4)产物提纯复杂; 5)制得的PLA易存在溶剂和催化剂残留,无法应用于医学领域[ |
开环聚合 Ring opening polymerization | 1)制得PLA的分子量高且集中; 2)制得PLA纯度高,力学性能好; 3)易调控PLA的化学结构,获得指定产物 | 1)丙交酯纯度要求高,纯化难度大; 2)合成工艺复杂、成本高、产率低 |
生物聚合 Biopolymerization | 1)反应条件温和,不易引入杂质和副反应; 2)PLA中不存在残留单体和毒性物质; 3)易生产优异对映纯度的聚合物; 4)易实现乳酸和其他单体的共聚 | 1)PLA产率低、分子量低; 2)生产成本和材料性能不可观; 3)尚处于实验室研究阶段,难以大规模应用 |
关键酶 Key enzyme | 来源菌株 Source strain | 宿主菌株 Host strain | 突变方式 Mutation method | 酶改造效果 Effect of enzyme modification | 参考文献 Reference | |
|---|---|---|---|---|---|---|
丙酰辅酶A转移酶 propionyl-CoA transferase | Clostridium ropionicum | E. coli XL1-Blue | V193A和4个沉默突变T78C、T669C、A1125G、T1158C(Pct540 Cp ) | 减缓酶表达对菌株的生长抑制,增强LA-CoA的合成,与野生型相比聚合物产量提高约5倍,聚合物中乳酸分数提高约8倍,酶体外特异性活性降低约39% | [ | |
| A243T和1个沉默突变A1200G(Pct532 Cp ) | 减缓酶表达对菌株的生长抑制,增强LA-CoA的合成,与野生型相比聚合物产量提高约4.5倍,聚合物中乳酸分数提高约9.5倍,酶体外特异性活性降低约52% | |||||
PHA合成酶 PHA synthetase | Ⅰ型 Type Ⅰ | Ralstonia eutropha | E. coli LS5218 | A510S | 赋予酶对LA-CoA的聚合能力,共聚物中乳酸分数最高可达26 mol%,且共聚物似为嵌段共聚物 | [ |
Ⅱ型 Type Ⅱ | Pseudomonas sp. 61-3 | E. coli JM109 | S325T/Q481K | 赋予酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至6 mol% | [ | |
| E. coli JW0885 | S325T/Q481K | 赋予酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至26 mol% | [ | |||
| S325T/Q481K/F392S | F392S的额外突变提高酶对LA-CoA的聚合能力,共聚物中乳酸分数提高约73%,聚合物含量提高约40.9% | |||||
| Pseudomonas sp. MBEL 6-19 | E. coli XL1-blue | E130D/Q481K | 赋予酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至35.3 mol% | [ | ||
| E130D/S477F/Q481K | 首次实现PLA均聚物(0.5 wt%)的合成,提高共聚物中乳酸分数至36.2 mol% | |||||
| E130D/S325T/Q481K | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至39.1 mol% | |||||
| E130D/S325T/S477R/Q481M | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至41.2 mol% | |||||
| E130D/S325T/S477F/Q481K | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至46 mol% | |||||
| E130D/S325T/S477G/Q481K | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至49 mol% | |||||
| Pseudomonas resinovorans | E. coli XL1-Blue | E130D/Q481K | 提高酶对LA-CoA的聚合能力,PLA产量提高至1.2 wt% | [ | ||
| E130D/S325T/S477G/Q481K | 提高酶对LA-CoA的聚合能力,PLA产量提高至7.3 wt% | |||||
嵌合酶 Chimerase | N端:Aeromonas cavia(Ⅱ型); C端:Ralstonia eutropha(Ⅰ型) | E. coli JM109 | N149D/F314H | 提高酶对LA-CoA的聚合能力, 共聚物中乳酸分数提高9.5倍 | [ | |
表2 聚乳酸生物合成关键酶的蛋白质工程策略与改造效果
Table 2 Protein engineering strategies and effects of key enzymes in PLA biosynthesis
关键酶 Key enzyme | 来源菌株 Source strain | 宿主菌株 Host strain | 突变方式 Mutation method | 酶改造效果 Effect of enzyme modification | 参考文献 Reference | |
|---|---|---|---|---|---|---|
丙酰辅酶A转移酶 propionyl-CoA transferase | Clostridium ropionicum | E. coli XL1-Blue | V193A和4个沉默突变T78C、T669C、A1125G、T1158C(Pct540 Cp ) | 减缓酶表达对菌株的生长抑制,增强LA-CoA的合成,与野生型相比聚合物产量提高约5倍,聚合物中乳酸分数提高约8倍,酶体外特异性活性降低约39% | [ | |
| A243T和1个沉默突变A1200G(Pct532 Cp ) | 减缓酶表达对菌株的生长抑制,增强LA-CoA的合成,与野生型相比聚合物产量提高约4.5倍,聚合物中乳酸分数提高约9.5倍,酶体外特异性活性降低约52% | |||||
PHA合成酶 PHA synthetase | Ⅰ型 Type Ⅰ | Ralstonia eutropha | E. coli LS5218 | A510S | 赋予酶对LA-CoA的聚合能力,共聚物中乳酸分数最高可达26 mol%,且共聚物似为嵌段共聚物 | [ |
Ⅱ型 Type Ⅱ | Pseudomonas sp. 61-3 | E. coli JM109 | S325T/Q481K | 赋予酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至6 mol% | [ | |
| E. coli JW0885 | S325T/Q481K | 赋予酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至26 mol% | [ | |||
| S325T/Q481K/F392S | F392S的额外突变提高酶对LA-CoA的聚合能力,共聚物中乳酸分数提高约73%,聚合物含量提高约40.9% | |||||
| Pseudomonas sp. MBEL 6-19 | E. coli XL1-blue | E130D/Q481K | 赋予酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至35.3 mol% | [ | ||
| E130D/S477F/Q481K | 首次实现PLA均聚物(0.5 wt%)的合成,提高共聚物中乳酸分数至36.2 mol% | |||||
| E130D/S325T/Q481K | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至39.1 mol% | |||||
| E130D/S325T/S477R/Q481M | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至41.2 mol% | |||||
| E130D/S325T/S477F/Q481K | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至46 mol% | |||||
| E130D/S325T/S477G/Q481K | 提高酶对LA-CoA的聚合能力, 提高共聚物中乳酸分数至49 mol% | |||||
| Pseudomonas resinovorans | E. coli XL1-Blue | E130D/Q481K | 提高酶对LA-CoA的聚合能力,PLA产量提高至1.2 wt% | [ | ||
| E130D/S325T/S477G/Q481K | 提高酶对LA-CoA的聚合能力,PLA产量提高至7.3 wt% | |||||
嵌合酶 Chimerase | N端:Aeromonas cavia(Ⅱ型); C端:Ralstonia eutropha(Ⅰ型) | E. coli JM109 | N149D/F314H | 提高酶对LA-CoA的聚合能力, 共聚物中乳酸分数提高9.5倍 | [ | |
碳源 Carbon source | 宿主菌株 Host strain | 使用的关键酶 Key enzyme | 代谢工程调控策略 Metabolic engineering regulation strategies | 产量 Yield | 分子量 Molecular weight | 参考文献 Reference |
|---|---|---|---|---|---|---|
葡萄糖 Glucose | E. coli XL1-blue | Pct540 Cp PhaC Ps6-19E130D/S325T/S477R/Q481M | 1)乳酸供应强化:过表达ldhA;敲除ppc 2)乙酰辅酶A供应强化:敲除ackA、adhE;过表达acs | 11 wt% | N.D. | [ |
| E. coli XL1-blue | Pct540 Cp PhaC Ps6-19E130D/S477F/Q481K | 1)启动子改造:敲除lacI 2)乳酸供应强化:替换ldhA的 天然启动子为强启动子trc 3)乙酰辅酶A供应强化:过表达acs 4)副产物阻断:敲除pflB、frdABCD和adhE | 4.2 wt% | 21 000 Da | [ | |
| E. coli BL21(DE3) | Pct540 Cp PhaC Cs | 形态工程:过表达sulA | 2.23 wt% (955 mg/L) | 21 000 Da | [ | |
| Corynebacterium glutamicum ATCC13803 | Pct Me PhaC1 PsSTQK | 聚合途径强化:过表达PhaC1 PsSTQK | 1.4 wt% | Mw: 5.7 kD; Mn: 4.3 kD | [ | |
| Yarrowia lipolytica | Pct540 Cp PhaC PaE130D/S325T/S477R/Q481M | 1)乳酸供应强化:敲除YlDLD1 2)聚合途径强化:过表达Pct540 Cp 、PhaC PaE130D/S325T/S477R/Q481M 3)区室化工程:细胞质表达Pct540 Cp,过氧化物酶体表达PhaC PaE130D/S325T/S477R/Q481M | 26 mg/g DCW | 50.5 kD | [ | |
二氧化碳 Carbon dioxide | Synechococcus elongatus PCC7942 | Pct540 Cp PhaC Ps6-19 | 1)聚合途径强化:过表达Pct540 Cp 、PhaC Ps6-19 2)乙酰辅酶A供给强化:下调ackA,过表达acs 3)脂肪酸途径弱化:下调accABCD、fabH、fabF 4)高密度发酵和培养条件优化 | 23 mg/g DCW (108 mg/L) | Mw: 62.5 kD;Mn: 32.8 kD | [ |
表3 生物合成聚乳酸的实例及使用的代谢工程调控策略
Table 3 Examples of biosynthesis of polylactic acid and its metabolic engineering regulation strategies
碳源 Carbon source | 宿主菌株 Host strain | 使用的关键酶 Key enzyme | 代谢工程调控策略 Metabolic engineering regulation strategies | 产量 Yield | 分子量 Molecular weight | 参考文献 Reference |
|---|---|---|---|---|---|---|
葡萄糖 Glucose | E. coli XL1-blue | Pct540 Cp PhaC Ps6-19E130D/S325T/S477R/Q481M | 1)乳酸供应强化:过表达ldhA;敲除ppc 2)乙酰辅酶A供应强化:敲除ackA、adhE;过表达acs | 11 wt% | N.D. | [ |
| E. coli XL1-blue | Pct540 Cp PhaC Ps6-19E130D/S477F/Q481K | 1)启动子改造:敲除lacI 2)乳酸供应强化:替换ldhA的 天然启动子为强启动子trc 3)乙酰辅酶A供应强化:过表达acs 4)副产物阻断:敲除pflB、frdABCD和adhE | 4.2 wt% | 21 000 Da | [ | |
| E. coli BL21(DE3) | Pct540 Cp PhaC Cs | 形态工程:过表达sulA | 2.23 wt% (955 mg/L) | 21 000 Da | [ | |
| Corynebacterium glutamicum ATCC13803 | Pct Me PhaC1 PsSTQK | 聚合途径强化:过表达PhaC1 PsSTQK | 1.4 wt% | Mw: 5.7 kD; Mn: 4.3 kD | [ | |
| Yarrowia lipolytica | Pct540 Cp PhaC PaE130D/S325T/S477R/Q481M | 1)乳酸供应强化:敲除YlDLD1 2)聚合途径强化:过表达Pct540 Cp 、PhaC PaE130D/S325T/S477R/Q481M 3)区室化工程:细胞质表达Pct540 Cp,过氧化物酶体表达PhaC PaE130D/S325T/S477R/Q481M | 26 mg/g DCW | 50.5 kD | [ | |
二氧化碳 Carbon dioxide | Synechococcus elongatus PCC7942 | Pct540 Cp PhaC Ps6-19 | 1)聚合途径强化:过表达Pct540 Cp 、PhaC Ps6-19 2)乙酰辅酶A供给强化:下调ackA,过表达acs 3)脂肪酸途径弱化:下调accABCD、fabH、fabF 4)高密度发酵和培养条件优化 | 23 mg/g DCW (108 mg/L) | Mw: 62.5 kD;Mn: 32.8 kD | [ |
| 1 | Stubbins A, Law KL, Muñoz SE, et al. Plastics in the earth system [J]. Science, 2021, 373(6550): 51-55. |
| 2 | MacLeod M, Arp HPH, Tekman MB, et al. The global threat from plastic pollution [J]. Science, 2021, 373(6550): 61-65. |
| 3 | Lau WWY, Shiran Y, Bailey RM, et al. Evaluating scenarios toward zero plastic pollution [J]. Science, 2020, 369(6510): 1455-1461. |
| 4 | Soo XYD, Jia LR, Lim QF, et al. Hydrolytic degradation and biodegradation of polylactic acid electrospun fibers [J]. Chemosphere, 2024, 350: 141186. |
| 5 | Ma YX, Guo XY, Du MM, et al. Beyond biodegradation: upcycling of polylactic acid plastic waste into amino acids via cascade catalysis under mild conditions [J]. Green Chem, 2024, 26(7): 3995-4004. |
| 6 | Hao LT, Ren SL, Li JY, et al. Feasibility of biodegradable material polylactic acid as a substitute for polypropylene for disposable medical masks production verified by life cycle assessment [J]. J Clean Prod, 2024, 448: 141492. |
| 7 | Yang TH, Jung YK, Kang HO, et al. Tailor-made type II Pseudomonas PHA synthases and their use for the biosynthesis of polylactic acid and its copolymer in recombinant Escherichia coli [J]. Appl Microbiol Biotechnol, 2011, 90(2): 603-614. |
| 8 | Han X, Liu JQ, Tian S, et al. Microbial cell factories for bio-based biodegradable plastics production [J]. iScience, 2022, 25(11): 105462. |
| 9 | Park SJ, Lee SY, Kim TW, et al. Biosynthesis of lactate-containing polyesters by metabolically engineered bacteria [J]. Biotechnol J, 2012, 7(2): 199-212. |
| 10 | Van Wouwe P, Dusselier M, Vanleeuw E, et al. Lactide synthesis and chirality control for polylactic acid production [J]. ChemSusChem, 2016, 9(9): 907-921. |
| 11 | Yang WJ, Zhou QK, Pan WH, et al. Synthesis of vanillin-based porphyrin for remarkably enhancing the toughness, UV-resistance and self-extinguishing properties of polylactic acid [J]. Chem Eng J, 2023, 469: 143935. |
| 12 | Hu X, Wang BT, Guo ZH, et al. Roles of phosphoramide derivatives in flame retardancy, thermal degradation and crystallization behaviors of polylactic acid [J]. Int J Biol Macromol, 2022, 219: 558-570. |
| 13 | Makri SP, Xanthopoulou E, Valera MA, et al. Poly (lactic acid) composites with lignin and nanolignin synthesized by in situ reactive processing [J]. Polymers, 2023, 15(10): 2386. |
| 14 | Boarino A, Schreier A, Leterrier Y, et al. Uniformly dispersed poly (lactic acid)-grafted lignin nanoparticles enhance antioxidant activity and UV-barrier properties of poly (lactic acid) packaging films [J]. ACS Appl Polym Mater, 2022, 4(7): 4808-4817. |
| 15 | Huang SY, Xue YF, Yu B, et al. A review of the recent developments in the bioproduction of polylactic acid and its precursors optically pure lactic acids [J]. Molecules, 2021, 26(21): 6446. |
| 16 | Gao CT, Wang YW, Yang YL, et al. Poly (lactic acid) synthesized from non-food biomass feedstocks with tin-loaded ZA molecular sieve catalysts by direct melt polycondensation [J]. Polym Int, 2024, 73(4): 310-318. |
| 17 | Wang QM, Chen XY, Zeng SH, et al. In-situ polycondensate-coated cellulose nanofiber heterostructure for polylactic acid-based composites with superior mechanical and thermal properties [J]. Int J Biol Macromol, 2023, 240: 124515. |
| 18 | van den Berg SA, Zuilhof H, Wennekes T. Clickable polylactic acids by fast organocatalytic ring-opening polymerization in continuous flow [J]. Macromolecules, 2016, 49(6): 2054-2062. |
| 19 | Stopper A, Okuda J, Kol M. Ring-opening polymerization of lactide with Zr complexes of {ONSO} ligands: from heterotactically inclined to isotactically inclined poly (lactic acid) [J]. Macromolecules, 2012, 45(2): 698-704. |
| 20 | Taguchi S, Yamada M, Matsumoto K, et al. A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme [J]. Proc Natl Acad Sci U S A, 2008, 105(45): 17323-17327. |
| 21 | Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic [J]. Microbiol Mol Biol Rev, 1999, 63(1): 21-53. |
| 22 | Yang TH, Kim TW, Kang HO, et al. Biosynthesis of polylactic acid and its copolymers using evolved propionate CoA transferase and PHA synthase [J]. Biotechnol Bioeng, 2010, 105(1): 150-160. |
| 23 | 谢彬, 白茸茸, 孙华山, 等. 聚乳酸塑料合成、生物降解及其废弃物处置的研究进展 [J]. 生物工程学报, 2023, 39(5): 1912-1929. |
| Xie B, Bai RR, Sun HS, et al. Synthesis, biodegradation and waste disposal of polylactic acid plastics: a review [J]. Chin J Biotechnol, 2023, 39(5): 1912-1929. | |
| 24 | Li G, Zhao MH, Xu F, et al. Synthesis and biological application of polylactic acid [J]. Molecules, 2020, 25(21): 5023. |
| 25 | Yu L, Zhao JB, Xu MM, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production: effects of CoA transferase [J]. Appl Microbiol Biotechnol, 2015, 99(11): 4917-4930. |
| 26 | Matsumoto K, Taguchi S. Biosynthetic polyesters consisting of 2-hydroxyalkanoic acids: current challenges and unresolved questions [J]. Appl Microbiol Biotechnol, 2013, 97(18): 8011-8021. |
| 27 | Volodina E, Schürmann M, Lindenkamp N, et al. Characterization of propionate CoA-transferase from Ralstonia eutropha H16 [J]. Appl Microbiol Biotechnol, 2014, 98(8): 3579-3589. |
| 28 | Matsumoto K, Taguchi S. Enzymatic and whole-cell synthesis of lactate-containing polyesters: toward the complete biological production of polylactate [J]. Appl Microbiol Biotechnol, 2010, 85(4): 921-932. |
| 29 | Lindenkamp N, Schürmann M, Steinbüchel A. A propionate CoA-transferase of Ralstonia eutropha H16 with broad substrate specificity catalyzing the CoA thioester formation of various carboxylic acids [J]. Appl Microbiol Biotechnol, 2013, 97(17): 7699-7709. |
| 30 | Park SJ, Kang KH, Lee H, et al. Propionyl-CoA dependent biosynthesis of 2-hydroxybutyrate containing polyhydroxyalkanoates in metabolically engineered Escherichia coli [J]. J Biotechnol, 2013, 165(2): 93-98. |
| 31 | Mezzolla V, D'Urso OF, Poltronieri P. Role of PhaC type I and type II enzymes during PHA biosynthesis [J]. Polymers, 2018, 10(8): 910. |
| 32 | Taguchi S. Current advances in microbial cell factories for lactate-based polyesters driven by lactate-polymerizing enzymes: towards the further creation of new LA-based polyesters [J]. Polym Degrad Stab, 2010, 95(8): 1421-1428. |
| 33 | Chek MF, Hiroe A, Hakoshima T, et al. PHA synthase (PhaC): interpreting the functions of bioplastic-producing enzyme from a structural perspective [J]. Appl Microbiol Biotechnol, 2019, 103(3): 1131-1141. |
| 34 | Zou HB, Shi MX, Zhang TT, et al. Natural and engineered polyhydroxyalkanoate (PHA) synthase: key enzyme in biopolyester production [J]. Appl Microbiol Biotechnol, 2017, 101(20): 7417-7426. |
| 35 | Tsuge T, Hyakutake M, Mizuno K. Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus [J]. Appl Microbiol Biotechnol, 2015, 99(15): 6231-6240. |
| 36 | Rehm BHA. Polyester synthases: natural catalysts for plastics [J]. Biochem J, 2003, 376(Pt 1): 15-33. |
| 37 | Yuan W, Jia Y, Tian J, et al. Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies [J]. Arch Biochem Biophys, 2001, 394(1): 87-98. |
| 38 | Li AP, Cao XP, Fu RZ, et al. Biocatalysis of CO2 and CH4: key enzymes and challenges [J]. Biotechnol Adv, 2024, 72: 108347. |
| 39 | Liu Q, Xun GH, Feng Y. The state-of-the-art strategies of protein engineering for enzyme stabilization [J]. Biotechnol Adv, 2019, 37(4): 530-537. |
| 40 | Ochi A, Matsumoto K, Ooba T, et al. Engineering of class I lactate-polymerizing polyhydroxyalkanoate synthases from Ralstonia eutropha that synthesize lactate-based polyester with a block nature [J]. Appl Microbiol Biotechnol, 2013, 97(8): 3441-3447. |
| 41 | Yamada M, Matsumoto K, Shimizu K, et al. Adjustable mutations in lactate (LA)-polymerizing enzyme for the microbial production of LA-based polyesters with tailor-made monomer composition [J]. Biomacromolecules, 2010, 11(3): 815-819. |
| 42 | Phan HT, Furukawa S, Imai K, et al. Biosynthesis of high-molecular-weight poly(d-lactate)-containing block copolyesters using evolved sequence-regulating polyhydroxyalkanoate synthase PhaCAR [J]. ACS Sustainable Chem Eng, 2023, 11(30): 11123-11129. |
| 43 | Selmer T, Willanzheimer A, Hetzel M. Propionate CoA-transferase from Clostridium propionicum. Cloning of gene and identification of glutamate 324 at the active site [J]. Eur J Biochem, 2002, 269(1): 372-380. |
| 44 | Shi MX, Li MD, Yang AR, et al. Class I polyhydroxyalkanoate (PHA) synthase increased polylactic acid production in engineered Escherichia coli [J]. Front Bioeng Biotechnol, 2022, 10: 919969. |
| 45 | Matsumoto K, Hori C, Fujii R, et al. Dynamic changes of intracellular monomer levels regulate block sequence of polyhydroxyalkanoates in engineered Escherichia coli [J]. Biomacromolecules, 2018, 19(2): 662-671. |
| 46 | Jung YK, Kim TY, Park SJ, et al. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers [J]. Biotechnol Bioeng, 2010, 105(1): 161-171. |
| 47 | Jung YK, Lee SY. Efficient production of polylactic acid and its copolymers by metabolically engineered Escherichia coli [J]. J Biotechnol, 2011, 151(1): 94-101. |
| 48 | Matsumoto K, Tobitani K, Aoki S, et al. Improved production of poly(lactic acid)-like polyester based on metabolite analysis to address the rate-limiting step [J]. AMB Express, 2014, 4(1): 83. |
| 49 | Lajus S, Dusséaux S, Verbeke J, et al. Engineering the yeast Yarrowia lipolytica for production of polylactic acid homopolymer [J]. Front Bioeng Biotechnol, 2020, 8: 954. |
| 50 | Tan CL, Tao F, Xu P. Direct carbon capture for the production of high-performance biodegradable plastics by cyanobacterial cell factories [J]. Green Chem, 2022, 24(11): 4470-4483. |
| 51 | 邢敏钰, 冉淦侨, 谭丹. 酿酒酵母中萜类化合物的生物合成与代谢调控研究进展 [J]. 生物工程学报, 2024, 40(6): 1661-1693. |
| Xing MY, Ran GQ, Tan D. Advances in the biosynthesis and metabolic regulation of terpenoids in Saccharomyces cerevisiae [J]. Chin J Biotechnol, 2024, 40(6): 1661-1693. | |
| 52 | Krivoruchko A, Zhang YM, Siewers V, et al. Microbial acetyl-CoA metabolism and metabolic engineering [J]. Metab Eng, 2015, 28: 28-42. |
| 53 | 陈心宇, 李梦怡, 陈国强. 聚羟基脂肪酸酯PHA代谢工程研究30年 [J]. 生物工程学报, 2021, 37(5): 1794-1811. |
| Chen XY, Li MY, Chen GQ. Thirty years of metabolic engineering for biosynthesis of polyhydroxyalkanoates [J]. Chin J Biotechnol, 2021, 37(5): 1794-1811. | |
| 54 | Zheng YK, Cheng FY, Zheng B, et al. Enhancing single-cell hyaluronic acid biosynthesis by microbial morphology engineering [J]. Synth Syst Biotechnol, 2020, 5(4): 316-323. |
| 55 | Jordan A, Chandler J, MacCready JS, et al. Engineering cyanobacterial cell morphology for enhanced recovery and processing of biomass [J]. Appl Environ Microbiol, 2017, 83(9): e00053-17. |
| 56 | Wan L, Zhu YY, Ke JT, et al. Compartmentalization of pathway sequential enzymes into synthetic protein compartments for metabolic flux optimization in Escherichia coli [J]. Metab Eng, 2024, 85: 167-179. |
| 57 | Bertacchi S, Jayaprakash P, Morrissey JP, et al. Interdependence between lignocellulosic biomasses, enzymatic hydrolysis and yeast cell factories in biorefineries [J]. Microb Biotechnol, 2022, 15(3): 985-995. |
| 58 | Prangemeier T, Wildner C, Françani AO, et al. Yeast cell segmentation in microstructured environments with deep learning [J]. Biosystems, 2022, 211: 104557. |
| [1] | 饶峻, 赵晨, 李端华, 廖豪, 黄加雨, 王辂. 自诱导策略在麦角硫因生物合成中的应用[J]. 生物技术通报, 2025, 41(1): 333-346. |
| [2] | 马小翔, 马泽源, 刘亚月, 周龙建, 和羿帆, 张翼. 仿突变生物合成调控对土曲霉C23-3次生代谢产物的影响[J]. 生物技术通报, 2024, 40(8): 275-287. |
| [3] | 聂祝欣, 郭瑾, 乔子洋, 李微薇, 张学燕, 刘春阳, 王静. 黑果枸杞不同发育时期果实花色苷合成的转录组分析[J]. 生物技术通报, 2024, 40(8): 106-117. |
| [4] | 沈真辉, 曹瑶, 杨林雷, 罗祥英, 子灵山, 陆青青, 李荣春. 金耳和毛韧革菌麦角硫因生物合成基因的克隆及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 259-272. |
| [5] | 何玙冰, 付振浩, 李仁瀚, 刘秀霞, 刘春立, 杨艳坤, 李业, 白仲虎. 利用代谢工程在酿酒酵母中高效合成2-萘乙醇[J]. 生物技术通报, 2024, 40(7): 99-107. |
| [6] | 胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44. |
| [7] | 张美玉, 赵玉斌, 王灵云, 宋元达, 赵新河, 任晓洁. 微藻破囊壶菌产功能性脂肪酸DHA研究进展[J]. 生物技术通报, 2024, 40(6): 81-94. |
| [8] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [9] | 刘玉萍, 张维阳, 章卫民, 叶伟, 李冬利. Phomopsis tersa FS441聚酮杂萜类化合物生物合成基因启动子的鉴定[J]. 生物技术通报, 2024, 40(12): 248-255. |
| [10] | 王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112. |
| [11] | 陈治民, 李翠, 韦继天, 李昕然, 刘峄, 郭强. 绿原酸生物合成调控及其应用研究进展[J]. 生物技术通报, 2024, 40(1): 57-71. |
| [12] | 何思成, 张紫瑗, 韩雨晴, 苗琳, 张翠英, 于爱群. 解脂耶氏酵母细胞工厂生产多不饱和脂肪酸的研究进展[J]. 生物技术通报, 2024, 40(1): 72-85. |
| [13] | 李亮, 徐姗姗, 姜艳军. 生物合成法生产麦角硫因的研究进展[J]. 生物技术通报, 2024, 40(1): 86-99. |
| [14] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [15] | 程亚楠, 张文聪, 周圆, 孙雪, 李玉, 李庆刚. 乳酸乳球菌生产2'-岩藻糖基乳糖的途径构建及发酵培养基优化[J]. 生物技术通报, 2023, 39(9): 84-96. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||