








生物技术通报 ›› 2025, Vol. 41 ›› Issue (8): 175-185.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0377
• 研究报告 • 上一篇
白雨果1( ), 李婉迪1, 梁建萍1,2, 石志勇1,2, 卢庚龙1, 刘红军1, 牛景萍1,2(
), 李婉迪1, 梁建萍1,2, 石志勇1,2, 卢庚龙1, 刘红军1, 牛景萍1,2( )
)
收稿日期:2025-04-11
出版日期:2025-08-26
发布日期:2025-08-14
通讯作者:
牛景萍,女,博士,讲师,研究方向 :黄芪抗病和促生机理;E-mail: niujingping@sxau.edu.cn作者简介:白雨果,男,硕士研究生,研究方向 :黄芪生态种植;E-mail: 15837071231@163.com
基金资助:
BAI Yu-guo1( ), LI Wan-di1, LIANG Jian-ping1,2, SHI Zhi-yong1,2, LU Geng-long1, LIU Hong-jun1, NIU Jing-ping1,2(
), LI Wan-di1, LIANG Jian-ping1,2, SHI Zhi-yong1,2, LU Geng-long1, LIU Hong-jun1, NIU Jing-ping1,2( )
)
Received:2025-04-11
Published:2025-08-26
Online:2025-08-14
摘要:
目的 探究哈茨木霉T9131对黄芪幼苗促生的调控机制,为T9131提高黄芪产量提供理论依据。 方法 对生长30 d的黄芪幼苗进行哈茨木霉T9131孢子悬浮液灌根处理,分别在灌根后的0 h、24 h、48 h和45 d对黄芪幼苗根部进行取样,并在45 d测定黄芪幼苗的株高、根长和植株鲜重。通过RNA-Seq构建3个时间点0、24和48 h的转录组文库,利用DESeq2软件获得3个比较组(T_24 h vs T_0 h、T_48 h vs T_0 h、T_48 h vs T_24 h)中的差异表达基因(DEGs),对3组DEGs进行韦恩分析,明确3组共有DEGs;对共有DEGs进行KEGG富集分析筛选促生相关基因,并通过RT-qPCR分析T9131诱导45 d和外源激素ABA诱导初期黄芪幼苗促生相关基因的表达情况;对0 h、48 h和45 d的根部样品通过LC-MS/MS方法进行15种激素含量测定。 结果 T9131能显著促进黄芪幼苗的株高和鲜重。T9131诱导黄芪幼苗转录组分析表明,3个比较组中共有差异表达基因149个。KEGG富集通路分析表明,与促生相关的通路主要有萜类骨架生物合成、类胡萝卜素生物合成、二萜生物合成、氰基氨基酸代谢、玉米素生物合成和植物激素信号转导;通路中富集的基因类型有HMGR、CYP707A2、GA2ox2、E3.2.1.21、UGT73C6、PYL4、GH3.1和JAZ,共包括14个基因,除基因UGT73C6(DN24891_c1_g1)受T9131诱导下调表达外,其余基因均上调表达;在T9131诱导黄芪幼苗45 d时HMGR(DN10463_c0_g2、DN989_c0_g1、DN113450_c0_g1、DN989_c2_g1)、CYP707A2和JAZ等7个基因显著上调表达,其余基因均下调表达。14个基因受ABA诱导表达分析表明,HMGR(DN10463_c0_g2、DN989_c0_g1、DN113450_c0_g1、DN989_c2_g1)、CYP707A2(DN397_c0_g2)和E3.2.1.21(DN2073_c0_g1)等6个基因受ABA诱导表达显著下调,其余基因均上调表达。激素测定表明,T9131能显著降低ABA含量。 结论 哈茨木霉T9131可能通过调控黄芪促生相关基因HMGR、CYP707A2和JAZ的上调表达和降低ABA含量促进黄芪生长;外源ABA能降低HMGR和CYP707A2的表达。
白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185.
BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings[J]. Biotechnology Bulletin, 2025, 41(8): 175-185.
| 基因名称 Gene name | 基因序号 Gene_ID | 引物序列 Sequence (5′‒3′) | |
|---|---|---|---|
| HMGR | DN10463_c0_g2 | F: CACTGCACGGACAACTTCC | R: ATTGCCGACCAGAAACCC |
| DN989_c0_g1 | F: GGATTCCCAATCAGTGCC | R: ATTCGTCTTCGTCGTAGTAGGT | |
| DN113450_c0_g1 | F: AGTGGCTGGTTCTGTTCTCG | R: TGTGGCGTTATCGTAGGGA | |
| DN52213_c0_g1 | F: AAGGATTCCCAATCAGTGCC | R: CGAAGAAGATGACGAGGTAGATG | |
| DN989_c2_g1 | F: TTTCCCTTCTCGGATTTCA | R: AAGACCGCGTTGGTCACGT | |
| CYP707A2 | DN397_c0_g1 | F: CTCATAGCCTTGTGGAATAGTGT | R: AGAGCGTTTGAAGCAGTGTTA |
| DN397_c0_g2 | F: ACTGGCAGTAGTGTCACGAGC | R: CAAGGAGGTGTATGAAGCAAAA | |
| GA2ox2 | DN7261_c0_g1 | F: TGAACCACCGTGTACCAAAG | R: ATTCCAGGTCACCCATATCAC |
| E3.2.1.21 | DN19827_c0_g1 | F: AGCAAGAAGTTGGTAGTGGGTA | R: GATTAGAAGATGAGTATGGTGGC |
| DN2073_c0_g1 | F: AAGAACATCGACCTGGTGC | R: ACTTTAGGGATTATGCGGAAC | |
| UGT73C6 | DN24891_c1_g1 | F: CCAGGGTAGTGGGTAGGAGT | R: GAAGACGTACAAGCGGGAG |
| PYL4 | DN9921_c0_g2 | F: TAACTAACGCCATAGCAAACG | R: ACTTCACGAAGCGATCCAA |
| GH3.1 | DN11968_c0_g1 | F: TCCATTAGGTCCACCAGCAT | R: TGTACGGCATCGGCATTT |
| JAZ | DN64262_c0_g1 | F: CCAGGAACCGATGAAGTGA | R: ACCCTACAAAGGCAGCAGA |
| Am18S | F: TGCAGAATCCCGTGAACCATC | R: AGGCATCGGGCAACGATATG | |
表1 RT-qPCR引物
Table 1 RT-qPCR primers
| 基因名称 Gene name | 基因序号 Gene_ID | 引物序列 Sequence (5′‒3′) | |
|---|---|---|---|
| HMGR | DN10463_c0_g2 | F: CACTGCACGGACAACTTCC | R: ATTGCCGACCAGAAACCC |
| DN989_c0_g1 | F: GGATTCCCAATCAGTGCC | R: ATTCGTCTTCGTCGTAGTAGGT | |
| DN113450_c0_g1 | F: AGTGGCTGGTTCTGTTCTCG | R: TGTGGCGTTATCGTAGGGA | |
| DN52213_c0_g1 | F: AAGGATTCCCAATCAGTGCC | R: CGAAGAAGATGACGAGGTAGATG | |
| DN989_c2_g1 | F: TTTCCCTTCTCGGATTTCA | R: AAGACCGCGTTGGTCACGT | |
| CYP707A2 | DN397_c0_g1 | F: CTCATAGCCTTGTGGAATAGTGT | R: AGAGCGTTTGAAGCAGTGTTA |
| DN397_c0_g2 | F: ACTGGCAGTAGTGTCACGAGC | R: CAAGGAGGTGTATGAAGCAAAA | |
| GA2ox2 | DN7261_c0_g1 | F: TGAACCACCGTGTACCAAAG | R: ATTCCAGGTCACCCATATCAC |
| E3.2.1.21 | DN19827_c0_g1 | F: AGCAAGAAGTTGGTAGTGGGTA | R: GATTAGAAGATGAGTATGGTGGC |
| DN2073_c0_g1 | F: AAGAACATCGACCTGGTGC | R: ACTTTAGGGATTATGCGGAAC | |
| UGT73C6 | DN24891_c1_g1 | F: CCAGGGTAGTGGGTAGGAGT | R: GAAGACGTACAAGCGGGAG |
| PYL4 | DN9921_c0_g2 | F: TAACTAACGCCATAGCAAACG | R: ACTTCACGAAGCGATCCAA |
| GH3.1 | DN11968_c0_g1 | F: TCCATTAGGTCCACCAGCAT | R: TGTACGGCATCGGCATTT |
| JAZ | DN64262_c0_g1 | F: CCAGGAACCGATGAAGTGA | R: ACCCTACAAAGGCAGCAGA |
| Am18S | F: TGCAGAATCCCGTGAACCATC | R: AGGCATCGGGCAACGATATG | |
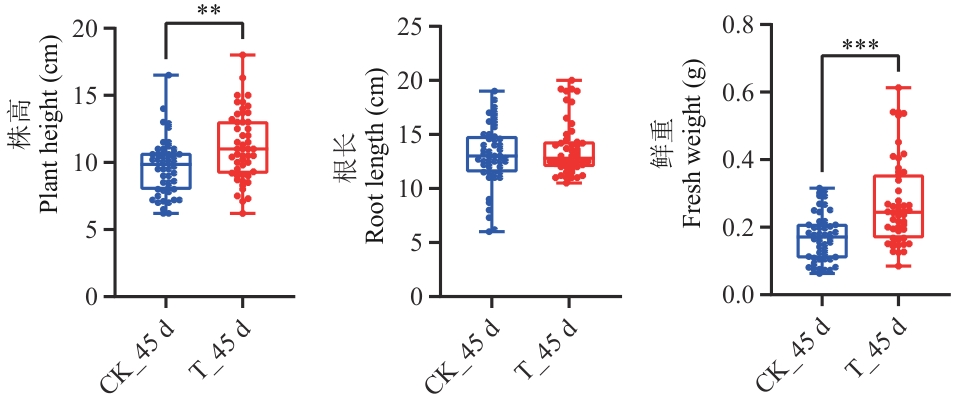
图1 哈茨木霉T9131对黄芪幼苗生长的作用CK表示用ddH2O灌根处理后的黄芪幼苗,T表示用T9131灌根处理后的黄芪幼苗;下划线后数字表示处理时间;* P<0.05,** P<0.01,*** P<0.001。下同
Fig. 1 Effect of Trichoderma harzianum T9131 on the growth of A. membranaceus seedlingsCK refers to the A. membranaceus seedlings after being watered with ddH2O, T refers to the A. membranaceus seedlings after being watered with T9131. The numbers following the underline indicate treatment time. * P<0.05, ** P<0.01, and *** P<0.001. The same below
样品名称 Sample | 过滤后的reads数 Clean reads | 过滤后的总数据量 Clean bases | Q20 (%) | Q30 (%) | GC含量比例 GC content (%) |
|---|---|---|---|---|---|
| T_0 h_1 | 49 844 534 | 7 453 178 401 | 98.27 | 94.75 | 42.15 |
| T_0 h_2 | 41 238 544 | 6 167 259 149 | 98.19 | 94.54 | 42.31 |
| T_0 h_3 | 53 377 736 | 7 964 860 750 | 98.31 | 94.88 | 42.28 |
| T_24 h_1 | 42 676 268 | 6 312 001 975 | 98.27 | 94.77 | 42.34 |
| T_24 h_2 | 47 351 528 | 7 061 996 999 | 98.19 | 94.55 | 42.45 |
| T_24 h_3 | 51 754 072 | 7 733 095 911 | 98.26 | 94.75 | 42.63 |
| T_48 h_1 | 43 661 406 | 6 521 807 743 | 98.02 | 94.06 | 42.41 |
| T_48 h_2 | 50 643 772 | 7 548 606 160 | 98.30 | 94.86 | 42.29 |
| T_48 h_3 | 47 716 402 | 7 100 483 345 | 98.29 | 94.82 | 42.56 |
表2 测序数据统计结果
Table 2 Statistics results of sequencing results
样品名称 Sample | 过滤后的reads数 Clean reads | 过滤后的总数据量 Clean bases | Q20 (%) | Q30 (%) | GC含量比例 GC content (%) |
|---|---|---|---|---|---|
| T_0 h_1 | 49 844 534 | 7 453 178 401 | 98.27 | 94.75 | 42.15 |
| T_0 h_2 | 41 238 544 | 6 167 259 149 | 98.19 | 94.54 | 42.31 |
| T_0 h_3 | 53 377 736 | 7 964 860 750 | 98.31 | 94.88 | 42.28 |
| T_24 h_1 | 42 676 268 | 6 312 001 975 | 98.27 | 94.77 | 42.34 |
| T_24 h_2 | 47 351 528 | 7 061 996 999 | 98.19 | 94.55 | 42.45 |
| T_24 h_3 | 51 754 072 | 7 733 095 911 | 98.26 | 94.75 | 42.63 |
| T_48 h_1 | 43 661 406 | 6 521 807 743 | 98.02 | 94.06 | 42.41 |
| T_48 h_2 | 50 643 772 | 7 548 606 160 | 98.30 | 94.86 | 42.29 |
| T_48 h_3 | 47 716 402 | 7 100 483 345 | 98.29 | 94.82 | 42.56 |
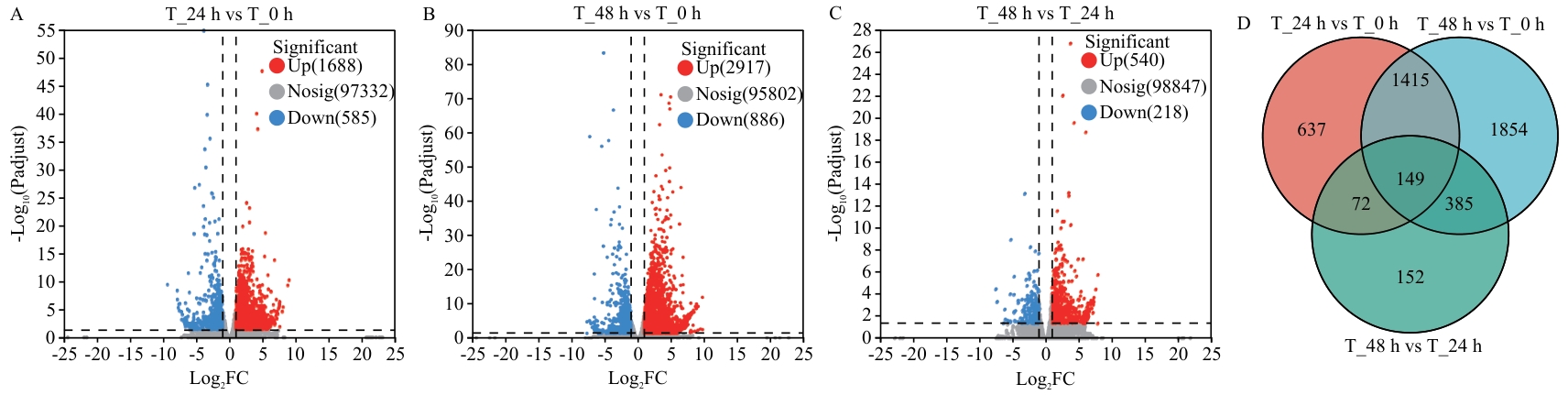
图3 黄芪幼苗受T9131诱导的差异基因分析(A-C)及韦恩分析(D)图A-C中,红色表示上调,蓝色表示下调,灰色指差异不显著基因
Fig. 3 DEGs analysis (A-C) and Venn analysis (D) of A. membranaceus seedlings by T9131 inducingIn Fig. A-C, red indicates upregulation, and blue indicates downregulation, and grey refers to genes with no significant difference

图5 差异表达基因KEGG富集分析横坐标指通路中富集的差异基因占总差异基因的比例,纵坐标指富集的KEGG通路
Fig. 5 KEGG enrichment analysis of DEGsThe x-axis refers to the ratio of the number of differentially expressed genes (DEGs) in the pathway to all DEGs; the y-axis refers to the KEGG pathway
KEGG通路 KEGG pathway | 基因名称 Gene name | 基因功能描述 Gene function description | 基因ID Gene_ID | T_24 h vs T_0 h (log2FC) | T_48 h vs T_0 h (log2FC) | T_48 h vs T_24 h (log2FC) |
|---|---|---|---|---|---|---|
萜类骨架生物合成 Terpenoid backbone biosynthesis (map00900) | HMGR | 3-羟基-3-甲基戊二酰辅酶A还原酶 | DN10463_c0_g2 | 2.129 9 | 3.611 3 | 1.457 1 |
| DN989_c0_g1 | 1.904 1 | 3.338 9 | 1.409 5 | |||
| DN113450_c0_g1 | 1.599 3 | 3.385 2 | 1.771 3 | |||
| DN52213_c0_g1 | 2.369 7 | 3.898 2 | 1.497 5 | |||
| DN989_c2_g1 | 2.061 1 | 3.914 2 | 1.825 3 | |||
类胡萝卜素生物合成 Carotenoid biosynthesis (map00906) | CYP707A2 | ABA 8'-羟化酶编码基因 | DN397_c0_g2 | 1.825 3 | 4.789 2 | 1.597 6 |
| DN397_c0_g1 | 2.256 5 | 3.766 6 | 1.482 3 | |||
二萜生物合成 Diterpenoid biosynthesis (map00904) | GA2ox2 | 赤霉素2-氧化酶 | DN7261_c0_g1 | 1.871 7 | 4.344 5 | 2.441 8 |
氰基氨基酸代谢 Cyanoamino acid metabolism (map00460) | E3.2.1.21 | β-葡萄糖苷酶 | DN19827_c0_g1 | 2.245 8 | 3.315 2 | 1.043 4 |
| DN2073_c0_g1 | 2.346 5 | 3.672 8 | 1.298 6 | |||
玉米素生物合成 Zeatin biosynthesis (map00908) | UGT73C6 | UDP-糖基转移酶 | DN24891_c1_g1 | -1.356 9 | -2.907 5 | -1.565 5 |
植物激素信号转导 Plant hormone signal transduction (map04075) | PYL4 | ABA受体 | DN9921_c0_g2 | 2.107 7 | 3.822 5 | 1.698 2 |
| GH3.1 | 吲哚-3-乙酸酰胺合成酶 | DN11968_c0_g1 | 1.889 8 | 3.488 6 | 1.571 7 | |
| JAZ | TIFY10A蛋白 | DN64262_c0_g1 | 2.062 9 | 3.588 1 | 1.499 8 |
表3 哈茨木霉T9131对黄芪促生机制相关的差异表达基因及功能注释
Table 3 DEGs and functional description of T. harzianum T9131 related to growth promotion mechanism of A. membranaceus
KEGG通路 KEGG pathway | 基因名称 Gene name | 基因功能描述 Gene function description | 基因ID Gene_ID | T_24 h vs T_0 h (log2FC) | T_48 h vs T_0 h (log2FC) | T_48 h vs T_24 h (log2FC) |
|---|---|---|---|---|---|---|
萜类骨架生物合成 Terpenoid backbone biosynthesis (map00900) | HMGR | 3-羟基-3-甲基戊二酰辅酶A还原酶 | DN10463_c0_g2 | 2.129 9 | 3.611 3 | 1.457 1 |
| DN989_c0_g1 | 1.904 1 | 3.338 9 | 1.409 5 | |||
| DN113450_c0_g1 | 1.599 3 | 3.385 2 | 1.771 3 | |||
| DN52213_c0_g1 | 2.369 7 | 3.898 2 | 1.497 5 | |||
| DN989_c2_g1 | 2.061 1 | 3.914 2 | 1.825 3 | |||
类胡萝卜素生物合成 Carotenoid biosynthesis (map00906) | CYP707A2 | ABA 8'-羟化酶编码基因 | DN397_c0_g2 | 1.825 3 | 4.789 2 | 1.597 6 |
| DN397_c0_g1 | 2.256 5 | 3.766 6 | 1.482 3 | |||
二萜生物合成 Diterpenoid biosynthesis (map00904) | GA2ox2 | 赤霉素2-氧化酶 | DN7261_c0_g1 | 1.871 7 | 4.344 5 | 2.441 8 |
氰基氨基酸代谢 Cyanoamino acid metabolism (map00460) | E3.2.1.21 | β-葡萄糖苷酶 | DN19827_c0_g1 | 2.245 8 | 3.315 2 | 1.043 4 |
| DN2073_c0_g1 | 2.346 5 | 3.672 8 | 1.298 6 | |||
玉米素生物合成 Zeatin biosynthesis (map00908) | UGT73C6 | UDP-糖基转移酶 | DN24891_c1_g1 | -1.356 9 | -2.907 5 | -1.565 5 |
植物激素信号转导 Plant hormone signal transduction (map04075) | PYL4 | ABA受体 | DN9921_c0_g2 | 2.107 7 | 3.822 5 | 1.698 2 |
| GH3.1 | 吲哚-3-乙酸酰胺合成酶 | DN11968_c0_g1 | 1.889 8 | 3.488 6 | 1.571 7 | |
| JAZ | TIFY10A蛋白 | DN64262_c0_g1 | 2.062 9 | 3.588 1 | 1.499 8 |
| [1] | 李博, 耿刚. 黄芪的化学成分与药理作用研究进展 [J]. 中西医结合研究, 2022, 14(4): 262-264. |
| Li B, Geng G. Research progress on chemical constituents and pharmacological effects of Astragalus membranaceus [J]. Res Integr Tradit Chin West Med, 2022, 14(4): 262-264. | |
| [2] | 马莹莹, 关一鸣, 王秋霞, 等. 黄芪主要病害及防治措施研究进展 [J]. 特产研究, 2019, 41(4): 101-107. |
| Ma YY, Guan YM, Wang QX, et al. Research progress on main diseases and control measures of Astragalus membranaceus [J]. Spec Wild Econ Anim Plant Res, 2019, 41(4): 101-107. | |
| [3] | 杨智慧. “恒山黄芪” 仿野生种植技术推广探析 [J]. 中国农机装备, 2023(8): 56-59. |
| Yang ZH. Analysis on the promotion of the imitation wild planting technology of “Astragalus Mount Heng” [J]. China Agric Mach Equip, 2023(8): 56-59. | |
| [4] | 周易, 李新星, 邹宇轩, 等. 不同种植模式黄芪药材质量与综合效益评价 [J]. 中国现代中药, 2025, 27(2): 296-303. |
| Zhou Y, Li XX, Zou YX, et al. Quality and comprehensive benefits of Astragalus membranaceus var. mongholicus cultivated in different patterns [J]. Mod Chin Med, 2025, 27(2): 296-303. | |
| [5] | 李俊达. 黄芪根系微生物的促生作用及微生物制剂对黄芪生长的大田效果 [D]. 呼和浩特: 内蒙古农业大学, 2024. |
| Li JD. Growth-promoting effects of root microorganisms of Astragalus membranaceus and field effects of microbial preparations on Astragalus growth [D]. Hohhot: Inner Mongolia Agricultural University, 2024. | |
| [6] | 王丽春. “恒山黄芪” 仿野生种植技术要点 [J]. 农机市场, 2024(10): 85-86. |
| Wang LC. Key points of imitation wild planting technology of “Hengshan Huangqi” [J]. Agric Mach Mark, 2024(10): 85-86. | |
| [7] | 李婷, 王洪旭, 崔广禄, 等. 哈茨木霉在植物应用上的研究进展 [J]. 中国农学通报, 2023, 39(21): 57-61. |
| Li T, Wang HX, Cui GL, et al. Application of Trichoderma harzianum in plant: research progress [J]. Chin Agric Sci Bull, 2023, 39(21): 57-61. | |
| [8] | Pani S, Kumar A, Sharma A. Trichoderma harzianum: an overview [J]. Bull Env Pharmacol Life Sci, 2021, 10(6): 32-39. |
| [9] | Li M, Ren YF, He C, et al. Complementary effects of dark septate endophytes and Trichoderma strains on growth and active ingredient accumulation of Astragalus mongholicus under drought stress [J]. J Fungi, 2022, 8(9): 920. |
| [10] | Shi ZY, Guo X, Lei ZH, et al. Screening of high-efficiency nitrogen-fixing bacteria from the traditional Chinese medicine plant Astragalus mongolicus and its effect on plant growth promotion and bacterial communities in the rhizosphere [J]. BMC Microbiol, 2023, 23(1): 292. |
| [11] | Shi ZY, Guo YX, Wang YY, et al. Nitrogen-fixing bacteria promote growth and bioactive components accumulation of Astragalus mongholicus by regulating plant metabolism and rhizosphere microbiota [J]. BMC Microbiol, 2024, 24(1): 261. |
| [12] | 刘旭, 苗莉云, 刘婷, 等. 产碱菌YNR32041对黄芪幼苗的促生作用分析 [J]. 北方园艺, 2023(19): 100-105. |
| Liu X, Miao LY, Liu T, et al. Analysis of the growth-promoting function of Alcaligenes sp. YNR32041 on Astragalus membranaceus seedlings [J]. North Hortic, 2023(19): 100-105. | |
| [13] | 杨振宇, 王宝慧, 宋诗娟, 等. 固氮菌剂对黄芪生长、土壤养分及酶活性的影响 [J]. 山西农业科学, 2022, 50(6): 861-868. |
| Yang ZY, Wang BH, Song SJ, et al. Effects of nitrogen-fixing bacteria on growth of Astragalus membranaceus, soil nutrient, and enzyme activity [J]. J Shanxi Agric Sci, 2022, 50(6): 861-868. | |
| [14] | 常佳钰, 陈婕, 赵博荣, 等. 光合细菌菌剂对栽培黄芪质量及其根际土壤理化性质的影响 [J]. 中药材, 2023, 46(4): 829-834. |
| Chang JY, Chen J, Zhao BR, et al. Effects of photosynthetic bacteria on quality of cultivated Astragalus membranaceus and physicochemical properties of rhizosphere soil [J]. J Chin Med Mater, 2023, 46(4): 829-834. | |
| [15] | 樊良帅, 黄振斌, 晋小军, 等. 三种生物菌剂对黄芪根腐病防效及生长发育的影响 [J]. 农药学学报, 2025, 27(1): 160-170. |
| Fan LS, Huang ZB, Jin XJ, et al. Effects of three microbial agents on the control of Astragalus membranaceus var. mongholicus root rot and their influence growth and development [J]. Chin J Pestic Sci, 2025, 27(1): 160-170. | |
| [16] | Chen YY, Shen Y, Chen BY, et al. Comparative transcriptome analyses between resistant and susceptible varieties in response to soybean mosaic virus infection [J]. Agronomy, 2022, 12(8): 1785. |
| [17] | 牛景萍, 燕翔, 白雨果, 等. 哈茨木霉T9131鉴定及其拮抗病原菌和诱导黄芪抗病的作用分析 [J/OL]. 广西植物, 2025. . |
| Niu JP, Yan X, Bai YG, et al. Identification of Trichoderma harzianum T9131 and analysis of its effect on against pathogen and disease resistance of Astragalus membranaceus [J/OL]. Guihaia, 2025. . | |
| [18] | Niu JP, Yan X, Bai YG, et al. Integration of transcriptomics and wgcna to characterize Trichoderma harzianum-induced systemic resistance in Astragalus mongholicus for defense against Fusarium solani [J]. Genes, 2024, 15(9): 1180. |
| [19] | Yang CQ, Fang X, Wu XM, et al. Transcriptional regulation of plant secondary metabolism [J]. J Integr Plant Biol, 2012, 54(10): 703-712. |
| [20] | Li Y, Zhou CX, Yan XJ, et al. Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry [J]. J Sep Sci, 2016, 39(10): 1804-1813. |
| [21] | Hui WK, Wang Y, Yan SJ, et al. Simultaneous analysis of endogenous plant growth substances during floral sex differentiation in Jatropha curcas L. using HPLC-ESI-MS/MS [J]. Sci Hortic, 2018, 241: 209-217. |
| [22] | Šimura J, Antoniadi I, Široká J, et al. Plant hormonomics: multiple phytohormone profiling by targeted metabolomics [J]. Plant Physiol, 2018, 177(2): 476-489. |
| [23] | Zhou LJ, Xiao LT, Xue HW. Dynamic cytology and transcriptional regulation of rice Lamina joint development [J]. Plant Physiol, 2017, 174(3): 1728-1746. |
| [24] | Hermosa R, Viterbo A, Chet I, et al. Plant-beneficial effects of Trichoderma and of its genes [J]. Microbiology, 2012, 158(Pt 1): 17-25. |
| [25] | 王席桃. 植物促生长内生细菌BH46调控鱼腥草生长及黄酮类组分代谢累积 [D]. 贵阳: 贵州师范大学, 2024. |
| Wang XT. Plant growth-promoting endophyte bacteria BH46 regulates the growth and metabolic accumulation of flavonoids in Houttuynia cordata thunb. [D]. Guiyang: Guizhou Normal University, 2024. | |
| [26] | 王楚彪, 卢万鸿, 林彦, 等. 转录组测序的发展和应用 [J]. 桉树科技, 2018, 35(4): 20-26. |
| Wang CB, Lu WH, Lin Y, et al. Development and application of transcriptome sequencing [J]. Eucalypt Sci Technol, 2018, 35(4): 20-26. | |
| [27] | Singh S, Chhatwal H, Pandey A. Deciphering the complexity of terpenoid biosynthesis and its multi-level regulatory mechanism in plants [J]. J Plant Growth Regul, 2024, 43(10): 3320-3336. |
| [28] | Bergman ME, Kortbeek RWJ, Gutensohn M, et al. Plant terpenoid biosynthetic network and its multiple layers of regulation [J]. Prog Lipid Res, 2024, 95: 101287. |
| [29] | Istvan ES, Deisenhofer J. The structure of the catalytic portion of human HMG-CoA reductase [J]. Biochim Biophys Acta Mol Cell Biol Lipds, 2000, 1529(1/2/3): 9-18. |
| [30] | Kushwaha RK, Singh S, Pandey SS, et al. Compatibility of inherent fungal endophytes of withania somnifera with Trichoderma viride and its impact on plant growth and withanolide content [J]. J Plant Growth Regul, 2019, 38(4): 1228-1242. |
| [31] | Schliemann W. Hydrolysis of conjugated gibberellins by β-glucosidases from dwarf rice (Oryza sativa L. cv. «Tan-ginbozu») [J]. J Plant Physiol, 1984, 116(2): 123-132. |
| [32] | Wuddineh WA, Mazarei M, Zhang JY, et al. Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance [J]. Plant Biotechnol J, 2015, 13(5): 636-647. |
| [33] | Zhou CW, Zheng YL, Leng J, et al. Effect of exogenous cadaverine phosphate on plant growth, photosynthesis, and gene expression of lettuce seedlings [J]. J Soil Sci Plant Nutr, 2024, 24(1): 537-546. |
| [34] | Ahmed W, Wang Y, Ji WX, et al. Unraveling the mechanism of the endophytic bacterial strain Pseudomonas oryzihabitans GDW1 in enhancing tomato plant growth through modulation of the host transcriptome and bacteriome [J]. Int J Mol Sci, 2025, 26(5): 1922. |
| [35] | Li C, Zheng LL, Wang XN, et al. Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights [J]. Plant Sci, 2019, 285: 1-13. |
| [36] | Cutler AJ, Krochko JE. Formation and breakdown of ABA [J]. Trends Plant Sci, 1999, 4(12): 472-478. |
| [37] | Bao DF, Chang SQ, Li XD, et al. Advances in the study of auxin early response genes: Aux/IAA GH3 and SAUR [J]. Crop J, 2024, 12(4): 964-978. |
| [38] | Liao DH, Chen X, Chen AQ, et al. The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis [J]. Plant Cell Physiol, 2015, 56(4): 674-687. |
| [39] | Han X, Kui MY, He KR, et al. Jasmonate-regulated root growth inhibition and root hair elongation [J]. J Exp Bot, 2023, 74(4): 1176-1185. |
| [40] | 王柯燕, 唐丽媚, 吴林捷, 等. 药用植物雷公藤JAZ基因家族的全基因组鉴定及功能分析 [J]. 药学学报, 2025, 60(4): 1156-1165. |
| Wang KY, Tang LM, Wu LJ, et al. Genome-wide identification and functional analysis of the JAZ gene family in Tripterygium wilfordii [J]. Acta Pharm Sin, 2025, 60(4): 1156-1165. | |
| [41] | 刘馨蕾. 蓝莓糖基转移酶基因家族鉴定及初步功能验证 [D]. 长春: 吉林农业大学, 2024. |
| Liu XL. Identification and preliminary functional validation of the UDP-glycosyltransferase gene family in blueberry [D]. Changchun: Jilin Agricultural University, 2024. |
| [1] | 任睿斌, 司二静, 万广有, 汪军成, 姚立蓉, 张宏, 马小乐, 李葆春, 王化俊, 孟亚雄. 大麦条纹病菌GH17基因家族的鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 146-154. |
| [2] | 王月琛, 韩鑫骐, 魏文敏, 崔兆兰, 罗阳美, 陈鹏如, 王海岗, 刘龙龙, 张莉, 王纶. 黍稷落粒的生物学基础研究及落粒调控基因的鉴定[J]. 生物技术通报, 2025, 41(7): 164-171. |
| [3] | 张越, 毕钰, 慕雪男, 郑子薇, 王志刚, 徐伟慧. 小麦赤霉病拮抗菌JB7的生防特性[J]. 生物技术通报, 2025, 41(7): 261-271. |
| [4] | 李成花, 豆飞飞, 任毓昭, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 外施水杨酸对白粉菌侵染小麦的影响及白粉菌转录组分析[J]. 生物技术通报, 2025, 41(7): 272-280. |
| [5] | 李凯月, 邓晓霞, 殷缘, 杜亚彤, 徐元静, 王竞红, 于耸, 蔺吉祥. 蓖麻LEA基因家族的鉴定和铝胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 128-138. |
| [6] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [7] | 韩燚, 侯昌林, 唐露, 孙璐, 谢晓东, 梁晨, 陈小强. 大麦HvERECTA基因的克隆及功能分析[J]. 生物技术通报, 2025, 41(7): 106-116. |
| [8] | 郭秀娟, 冯宇, 吴瑞香, 王利琴, 杨建春. Ca2+处理对胡麻种子萌发影响的转录组分析[J]. 生物技术通报, 2025, 41(7): 139-149. |
| [9] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [10] | 王苗苗, 赵相龙, 王召明, 刘志鹏, 闫龙凤. 花苜蓿TCP基因家族的鉴定及其在干旱胁迫下的表达模式分析[J]. 生物技术通报, 2025, 41(6): 179-190. |
| [11] | 瞿美玲, 周思敏, 张惊宇, 何佳蔚, 朱佳源, 刘笑蓉, 童巧珍, 周日宝, 刘湘丹. 灰毡毛忍冬bHLH转录基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(6): 256-268. |
| [12] | 刘鑫, 王嘉雯, 李进伟, 牟策, 杨盼盼, 明军, 徐雷锋. 兰州百合三个LdBBXs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(5): 186-196. |
| [13] | 李志强, 罗正乾, 徐琳黎, 周国慧, 屈丝雨, 刘恩良, 顼东婷. 基于T2T基因组鉴定大豆R2R3-MYB基因家族及干旱和盐胁迫下的表达分析[J]. 生物技术通报, 2025, 41(5): 141-152. |
| [14] | 胡若群, 曾菁菁, 梁婉凤, 曹佳玉, 黄小苇, 梁晓英, 仇明月, 陈莹. 转录组和代谢组联合分析探究不同遮光条件下金线莲类胡萝卜素合成代谢机制[J]. 生物技术通报, 2025, 41(5): 231-243. |
| [15] | 赵婧, 郭茜, 李睿琦, 雷滢炀, 岳爱琴, 赵晋忠, 殷丛丛, 杜维俊, 牛景萍. 大豆GmGST基因簇基因序列分析及诱导表达分析[J]. 生物技术通报, 2025, 41(5): 129-140. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||