








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 254-261.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0015
Previous Articles Next Articles
ZHANG Hao1,2( ), HE Chang-sheng1,2, LI Yan-yan1,2, WANG Yong1,2(
), HE Chang-sheng1,2, LI Yan-yan1,2, WANG Yong1,2( ), ZHU Jiang-jiang2, Emu Quzhe3, LIN Ya-qiu1,2(
), ZHU Jiang-jiang2, Emu Quzhe3, LIN Ya-qiu1,2( )
)
Received:2022-01-05
Online:2022-10-26
Published:2022-11-11
Contact:
WANG Yong,LIN Ya-qiu
E-mail:zhanghao9235@163.com;wangyong010101@swun.cn;linyq1999@163.com
ZHANG Hao, HE Chang-sheng, LI Yan-yan, WANG Yong, ZHU Jiang-jiang, Emu Quzhe, LIN Ya-qiu. Regulation of miR-301b on Goat Intramuscular Adipocyte Differentiation[J]. Biotechnology Bulletin, 2022, 38(10): 254-261.
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| miR-301b mimics | CAGUGCAAUGAUAUUGUCAAAGC UUUGACAAUAUCAUUGCACUGUU |
| mimics NC | UUCUCCGAACGUGUCACGUTTACGUGACACG- UUCGGAGAATT |
| miR-301b Inhibitor | GCUUUGACAAUAUCAUUGCACUG |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
Table 1 Sequences of miRNA mimic and inhibitor
| 名称Name | 序列Sequence(5'-3') |
|---|---|
| miR-301b mimics | CAGUGCAAUGAUAUUGUCAAAGC UUUGACAAUAUCAUUGCACUGUU |
| mimics NC | UUCUCCGAACGUGUCACGUTTACGUGACACG- UUCGGAGAATT |
| miR-301b Inhibitor | GCUUUGACAAUAUCAUUGCACUG |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
| 基因 Gene/miRNA | 序列 Sequence(5'-3') | Tm/℃ | 产物长度 Product length/ bp | 登录号 GenBank No. |
|---|---|---|---|---|
| AP2 | TGAAGTCACTCCAGATGACAGGTGACACATTCCAGCACCAGC | 58 | 143 | NM_001285623.1 |
| CEBPα | CCGTGGACAAGAACAGCAACAGGCGGTCATTGTCACTGGT | 58 | 142 | XM_018062278.1 |
| CEBPβ | CAAGAAGACGGTGGACAAGCAACAAGTTCCGCAGGGTG | 66 | 204 | XM_018058020.1 |
| PPARγ | AAGCGTCAGGGTTCCACTATG GAACCTGATGGCGTTATGAGAC | 60 | 197 | NM_001285658.1 |
| Pref1 | CCGGCTTCATGGATAAGACCTGCCTCGCACTTGTTGAGGAA | 65 | 184 | KP686197.1 |
| SREBP1 | AAGTGGTGGGCCTCTCTGA GCAGGGGTTTCTCGGACT | 58 | 127 | NM_001285755.1 |
| KLF3 | CTCGGTGTCATACCCGTCTAATCCATCTGCATCCCGTGAGAC | 60 | 128 | KU041753.1 |
| UXT | GCAAGTGGATTTGGGCTGTAACATGGAGTCCTTGGTGAGGTTGT | 60 | 180 | XM_005700842.2 |
| U6 | TGGAACGCTTCACGAATTTGCGGGAACGATACAGAGAAGATTAGC | 60 | ||
| miR-301b | CTGGAGCAGTGCAATGATGAAAGTGCAGGGTCCGAGGT | 58 |
Table 2 Sequences information of specific primers
| 基因 Gene/miRNA | 序列 Sequence(5'-3') | Tm/℃ | 产物长度 Product length/ bp | 登录号 GenBank No. |
|---|---|---|---|---|
| AP2 | TGAAGTCACTCCAGATGACAGGTGACACATTCCAGCACCAGC | 58 | 143 | NM_001285623.1 |
| CEBPα | CCGTGGACAAGAACAGCAACAGGCGGTCATTGTCACTGGT | 58 | 142 | XM_018062278.1 |
| CEBPβ | CAAGAAGACGGTGGACAAGCAACAAGTTCCGCAGGGTG | 66 | 204 | XM_018058020.1 |
| PPARγ | AAGCGTCAGGGTTCCACTATG GAACCTGATGGCGTTATGAGAC | 60 | 197 | NM_001285658.1 |
| Pref1 | CCGGCTTCATGGATAAGACCTGCCTCGCACTTGTTGAGGAA | 65 | 184 | KP686197.1 |
| SREBP1 | AAGTGGTGGGCCTCTCTGA GCAGGGGTTTCTCGGACT | 58 | 127 | NM_001285755.1 |
| KLF3 | CTCGGTGTCATACCCGTCTAATCCATCTGCATCCCGTGAGAC | 60 | 128 | KU041753.1 |
| UXT | GCAAGTGGATTTGGGCTGTAACATGGAGTCCTTGGTGAGGTTGT | 60 | 180 | XM_005700842.2 |
| U6 | TGGAACGCTTCACGAATTTGCGGGAACGATACAGAGAAGATTAGC | 60 | ||
| miR-301b | CTGGAGCAGTGCAATGATGAAAGTGCAGGGTCCGAGGT | 58 |
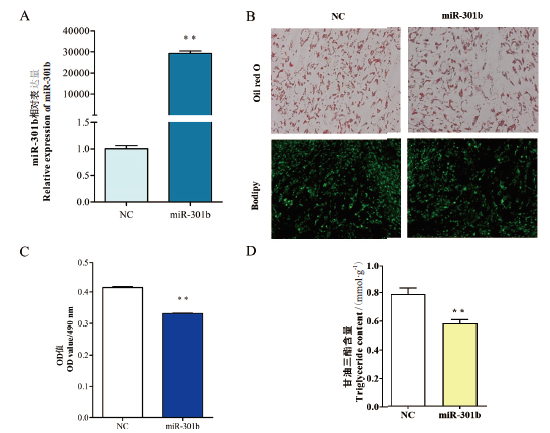
Fig.1 Effect of overexpressing miR-301b on the morphol-ogy of goat intramuscular adipocytes A:The overexpressing efficiency of miR-301b. B:Oil red O staining and Bodipy staining(×400). C:OD value of oil red O staining. D:Triglyceride(TG)content assay. * indicates P<0.05, ** indicates P<0.01, the same below
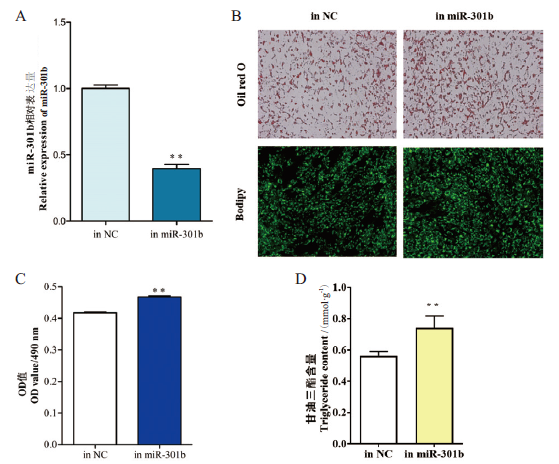
Fig. 3 Effect of interfering miR-301b on the morphology of goat intramuscular adipocytes A:Efficiency of miR-301b. B:Oil red O staining and Bodipy staining(×400).C:OD value of oil red O staining. D:Triglyceride(TG)content assay
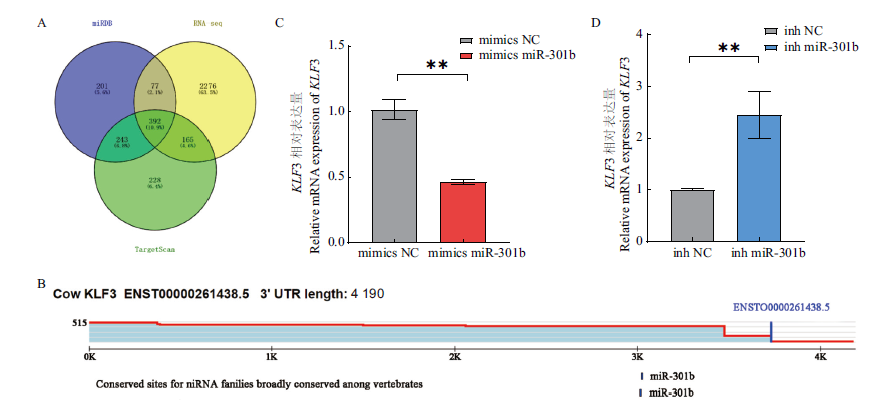
Fig. 5 Prediction and expression level detection of miR-301b target gene A:miR-301b target gene prediction Venn diagram;B:KLF3 potential miRNA binding site prediction;C:KLF3 expression level after simulating miR-301b expression;D:KLF3 expression level after interfering with miR-301b expression
| [1] |
Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation[J]. Physiol Rev, 1998, 78(3):783-809.
pmid: 9674695 |
| [2] | Liu HD, Li BJ, Qiao LY, et al. miR-340-5p inhibits sheep adipocyte differentiation by targeting ATF7[J]. Anim Sci J, 2020, 91(1):e13462. |
| [3] |
Wen ZY, Tang Z, Li MX, et al. APPL1 knockdown blocks adipogenic differentiation and promotes adipocyte lipolysis[J]. Mol Cell Endocrinol, 2020, 506:110755.
doi: 10.1016/j.mce.2020.110755 URL |
| [4] |
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene Lin-4 encodes small RNAs with antisense complementarity to Lin-14[J]. Cell, 1993, 75(5):843-854.
doi: 10.1016/0092-8674(93)90529-y pmid: 8252621 |
| [5] |
Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs[J]. Nature, 2004, 432(7014):235-240.
doi: 10.1038/nature03120 URL |
| [6] |
Ashrafizadeh M, Najafi M, Mohammadinejad R, et al. Flaming the fight against cancer cells:the role of microRNA-93[J]. Cancer Cell Int, 2020, 20:277.
doi: 10.1186/s12935-020-01349-x pmid: 32612456 |
| [7] |
Wang HY, Zheng Y, Wang GL, et al. Identification of microRNA and bioinformatics target gene analysis in beef cattle intramuscular fat and subcutaneous fat[J]. Mol Biosyst, 2013, 9(8):2154-2162.
doi: 10.1039/c3mb70084d pmid: 23728155 |
| [8] |
Ma XY, Wei DW, Cheng G, et al. Bta-miR-130a/b regulates preadipocyte differentiation by targeting PPARG and CYP2U1 in beef cattle[J]. Mol Cell Probes, 2018, 42:10-17.
doi: 10.1016/j.mcp.2018.10.002 URL |
| [9] | Xu HY, Shao J, Yin BZ, et al. Bovine bta-microRNA-1271 promotes preadipocyte differentiation by targeting activation transcription factor 3[J]. Biochemistry(Mosc), 2020, 85(7):749-757. |
| [10] | Yu X, Fang XB, Gao M, et al. Isolation and identification of bovine preadipocytes and screening of microRNAs associated with adipogenesis[J]. Animals(Basel), 2020, 10(5):818. |
| [11] |
Zhang M, Li DH, Li F, et al. Integrated analysis of MiRNA and genes associated with meat quality reveals that gga-miR-140-5p affects intramuscular fat deposition in chickens[J]. Cell Physiol Biochem, 2018, 46(6):2421-2433.
doi: 10.1159/000489649 pmid: 29742492 |
| [12] |
Wang WS, Du ZQ, Cheng BH, et al. Expression profiling of preadipocyte microRNAs by deep sequencing on chicken lines divergently selected for abdominal fatness[J]. PLoS One, 2015, 10(2):e0117843.
doi: 10.1371/journal.pone.0117843 URL |
| [13] | 靳文姣, 翟彬, 苑鹏涛, 等. miR-215-5p通过靶向NCOA3基因抑制固始鸡腹部前脂肪细胞的增殖和分化[J]. 畜牧与兽医, 2021, 53(7):69-77. |
| Jin WJ, Zhai B, Yuan PT, et al. miR-215-5p inhibits the proliferation and differentiation of Gushi chicken abdominal pre-adipocytes by targeting the NCOA3 gene[J]. Animal Husb Vet Med, 2021, 53(7):69-77. | |
| [14] | 何洪炳, 蔡明成, 梁小虎, 等. miR-130b靶向PPARγ抑制家兔前体脂肪细胞分化[J]. 畜牧兽医学报, 2017, 48(11):2076-2083. |
| He HB, Cai MC, Liang XH, et al. miR-130bInhibits the differentiation of rabbit preadipocytes by targeting PPARγ[J]. Chin J Animal Vet Sci, 2017, 48(11):2076-2083. | |
| [15] |
Li XF, He SS, Li RP, et al. Pseudomonas aeruginosa infection augments inflammation through miR-301b repression of c-Myb-mediated immune activation and infiltration[J]. Nat Microbiol, 2016, 1(10):16132.
doi: 10.1038/nmicrobiol.2016.132 URL |
| [16] | Peng Y, Xi X, Li JT, et al. miR-301b and NR3C2 co-regulate cells malignant properties and have the potential to be independent prognostic factors in breast cancer[J]. J Biochem Mol Toxicol, 2021, 35(2):e22650. |
| [17] |
Tatekoshi Y, Tanno M, Kouzu H, et al. Translational regulation by miR-301b upregulates AMP deaminase in diabetic hearts[J]. J Mol Cell Cardiol, 2018, 119:138-146.
doi: S0022-2828(18)30150-0 pmid: 29733818 |
| [18] |
Zhang L, Yang F, Yan Q. Candesartan ameliorates vascular smooth muscle cell proliferation via regulating miR-301b/STAT3 axis[J]. Hum Cell, 2020, 33(3):528-536.
doi: 10.1007/s13577-020-00333-x pmid: 32170715 |
| [19] |
Liu LL, Liu HH, Chen MT, et al. miR-301b-miR-130b-PPARγ axis underlies the adipogenic capacity of mesenchymal stem cells with different tissue origins[J]. Sci Rep, 2017, 7(1):1160.
doi: 10.1038/s41598-017-01294-2 URL |
| [20] | He CS, Wang Y, Xu Q, et al. Overexpression of Krueppel like factor 3 promotes subcutaneous adipocytes differentiation in goat Capra hircus[J]. Anim Sci J, 2021, 92(1):e13514. |
| [21] |
Rosen ED. The transcriptional basis of adipocyte development[J]. Prostaglandins Leukot Essent Fatty Acids, 2005, 73(1):31-34.
doi: 10.1016/j.plefa.2005.04.004 URL |
| [22] |
Chen Z. Progress and prospects of long noncoding RNAs in lipid homeostasis[J]. Mol Metab, 2015, 5(3):164-170.
doi: 10.1016/j.molmet.2015.12.003 URL |
| [23] |
McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity[J]. Curr Mol Med, 2011, 11(4):304-316.
doi: 10.2174/156652411795677990 pmid: 21506921 |
| [24] |
Ambros V. The functions of animal microRNAs[J]. Nature, 2004, 431(7006):350-355.
doi: 10.1038/nature02871 URL |
| [25] |
Ha MJ, Kim VN. Regulation of microRNA biogenesis[J]. Nat Rev Mol Cell Biol, 2014, 15(8):509-524.
doi: 10.1038/nrm3838 URL |
| [26] |
Bartel DP. microRNAs:genomics, biogenesis, mechanism, and function[J]. Cell, 2004, 116(2):281-297.
doi: 10.1016/s0092-8674(04)00045-5 pmid: 14744438 |
| [27] |
Lefterova MI, Zhang Y, Steger DJ, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale[J]. Genes Dev, 2008, 22(21):2941-2952.
doi: 10.1101/gad.1709008 URL |
| [28] |
Rosen ED, Hsu CH, Wang XZ, et al. C/EBPalpha induces adipogenesis through PPARgamma:a unified pathway[J]. Genes Dev, 2002, 16(1):22-26.
doi: 10.1101/gad.948702 URL |
| [29] |
Cirilli M, Bereshchenko O, Ermakova O, et al. Insights into specificity, redundancy and new cellular functions of C/EBPa and C/EBPb transcription factors through interactome network analysis[J]. Biochim Biophys Acta Gen Subj, 2017, 1861(2):467-476.
doi: 10.1016/j.bbagen.2016.10.002 URL |
| [30] | 柳晓峰, 李辉. PPAR基因与脂肪代谢调控[J]. 遗传, 2006, 28(2):243-248. |
| Liu XF, Li H. Regulation of fat metabolism by PPARs[J]. Hereditas, 2006, 28(2):243-248. | |
| [31] |
Liu FY, Zhang GL, Lv SM, et al. miRNA-301b-3p accelerates migration and invasion of high-grade ovarian serous tumor via targeting CPEB3/EGFR axis[J]. J Cell Biochem, 2019, 120(8):12618-12627.
doi: 10.1002/jcb.28528 pmid: 30834603 |
| [32] |
Pan SF, Yang XJ, Jia YM, et al. Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-g expression[J]. J Cell Physiol, 2014, 229(5):631-639.
doi: 10.1002/jcp.24486 pmid: 24311275 |
| [33] | 徐珂. 调控猪脂肪细胞代谢的关键microRNAs的鉴定[D]. 秦皇岛: 河北科技师范学院, 2020. |
| Xu K. Identification of key microRNAs regulating the metabolism of porcine adipocytes[D]. Qinhuangdao: Hebei Normal University of Science & Technology, 2020. | |
| [34] | 杜宇, 赵越, 林亚秋, 等. miR-106b-5p靶向KLF4调控山羊肌内前体脂肪细胞分化[J]. 畜牧兽医学报, 2020, 51(6):1219-1228. |
| Du Y, Zhao Y, Lin YQ, et al. miR-106b-5p regulates the differentiation of goat intramuscular preadipocytes by targeting KLF4[J]. Chin J Animal Vet Sci, 2020, 51(6):1219-1228. | |
| [35] | 王杰. miR-324-5p对猪脂肪沉积的作用研究及机制研究[D]. 杨凌: 西北农林科技大学, 2020. |
| Wang J. The effect and mechanism of miR-324-5p on pig fat deposition[D]. Yangling: Northwest A & F University, 2020. |
| [1] | LI Ying, YUE Xiang-hua. Application of DNA Methylation in Interpreting Natural Variation in Moso Bamboo [J]. Biotechnology Bulletin, 2023, 39(7): 48-55. |
| [2] | YANG Xin-ran, WANG Jian-fang, MA Xin-hao, ZAN Lin-sen. Expression Analyses of m6A Methylase Genes in Bovine Adipogenesis [J]. Biotechnology Bulletin, 2022, 38(7): 70-79. |
| [3] | SHENG Xue-qing, ZHAO Nan, LIN Ya-qiu, CHEN Ding-shuang, WANG Rui-long, LI Ao, WANG Yong, LI Yan-yan. Cloning and Expression Analysis of ZNF32 Gene in Goat [J]. Biotechnology Bulletin, 2022, 38(12): 300-311. |
| [4] | ZHU Wen, TANG Ying-ying, SUN Xin-yang, ZHOU Ming, ZHANG Zi-jun, CHEN Xing-yong. Effect of Low Crude Protein Diet on the Liver Transcriptome Sequencing of Goats [J]. Biotechnology Bulletin, 2021, 37(9): 203-211. |
| [5] | JIN Qiu-xia, WANG Si-hong, JIN Li-hua. Research Progress on Drosophila Intestinal Stem Cells and Intestinal Microflora [J]. Biotechnology Bulletin, 2021, 37(4): 245-250. |
| [6] | ZHANG Hao, ZHANG Ya-nan, LI Xin, WANG Jia-mei, WANG Yong, ZHU Jiang-jiang, XIONG Yan, LIN Ya-qiu. Effect of PDK4 on the Lipid Metabolism of Goat Intramuscular Adipocytes [J]. Biotechnology Bulletin, 2021, 37(12): 151-159. |
| [7] | ZHANG Le-chao, LIU Yue-qin, DUAN Chun-hui, ZHANG Ying-jie, WANG Yong, GUO Yun-xia. Analysis of Genetic Diversity and Genetic Structure in 7 Local Goat Breeds [J]. Biotechnology Bulletin, 2020, 36(6): 183-190. |
| [8] | LI Xiao-kai, FAN Yi-xing, QIAO Xian, ZHANG Lei, WANG Feng-hong, WANG Zhi-ying, WANG Rui-jun, ZHANG Yan-jun, LIU Zhi-hong, WANG Zhi-xin, HE Li-bing, LI Jin-quan, SU Rui, ZHANG Jia-xin. Research Progress of Goat Genome and Genetic Variation Map [J]. Biotechnology Bulletin, 2020, 36(4): 175-184. |
| [9] | SONG Shao-zheng, LU Rui, ZHANG Ting, HE Zheng-yi, WU Zhao-manqiu, CHENG Yong, ZHOU Ming-ming. Research Progress of CRISPR /Cas9 Gene Editing Technology in Goat and Sheep [J]. Biotechnology Bulletin, 2020, 36(3): 62-68. |
| [10] | LI Kun, LIU Yue, HUANG Peng, YANG Zhi-fang, HU Qian, ZHANG Ying, LI Zhi-hong, LÜ Ye-hui, LIANG Le. Proteomics Study on Spermatogonia Differentiation in Mice [J]. Biotechnology Bulletin, 2020, 36(3): 168-176. |
| [11] | WANG Hong-fang, XU Bao-hua. The Relationship Between Gut Microflora and Sociality of Honeybee [J]. Biotechnology Bulletin, 2020, 36(2): 71-76. |
| [12] | ZHOU Min-ya, LU Rui, ZHANG Ting, YUAN Ting-ting, LU Yao-yao, YAN Kun-ning, YUAN Yu-guo, CHENG Yong. Preparation of Recombinant Human SOD1/3 Transgenic Goat and Detection of Expressed Products [J]. Biotechnology Bulletin, 2019, 35(5): 85-92. |
| [13] | HUANG Long, WU Ben-li, HE Ji-xiang, CHEN Jing, SONG Guang-tong, WANG Xiang, ZHANG Ye, WU Song. SNP Identification of MyoD1 Gene and Its Correlation with Growth Traits in Pelodiscus sinensis [J]. Biotechnology Bulletin, 2019, 35(4): 76-81. |
| [14] | DAI Feng-bin, LIU Li-ping, LI Ai-jia, RAO Shu-pei, CHEN Jin-huan. Establishment of Highly Efficient and Rapid Propagation System of Lycium ruthenicum for Multiple Genotypes [J]. Biotechnology Bulletin, 2019, 35(4): 201-207. |
| [15] | GAO Jie ,TONG Shuo-qiu ,ZHONG Jie ,GONG Xian, WU Yong-jun. Cloning of a HD-ZIP Transcription Factor Gene HD20 and A Preliminary Study on Function of Fiber Differentiation in Tobacco [J]. Biotechnology Bulletin, 2018, 34(6): 84-89. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||