








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 305-314.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0384
Previous Articles Next Articles
HAN Xue1,2( ), ZHANG A-na3, WANG Hai-yan4, XIN Feng-jiao1,2, GU Tian-yi1,2(
), ZHANG A-na3, WANG Hai-yan4, XIN Feng-jiao1,2, GU Tian-yi1,2( ), WANG Yu-lu1,2(
), WANG Yu-lu1,2( )
)
Received:2024-04-15
Online:2024-10-26
Published:2024-11-20
Contact:
GU Tian-yi, WANG Yu-lu
E-mail:njhanxue1995@163.com;18501151081@163.com;wnewyx@163.com
HAN Xue, ZHANG A-na, WANG Hai-yan, XIN Feng-jiao, GU Tian-yi, WANG Yu-lu. Computer-aided Thermostability Engineering and Underlying Mechanism Investigation of the GH11 Family Xylanase CDBFV[J]. Biotechnology Bulletin, 2024, 40(10): 305-314.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| N37P-F | CGGATAGCGGCCCGAATAGCGCGACCTTTTATA-G |
| N37P-R | CGCGCTATTCGGGCCGCTATCCGCCCACAGTTCATAG |
| N38V-F | CGGATAGCGGCAATGTTAGCGCGACCTTTTATAGCGATGGC |
| N38V-R | GGTCGCGCTGTTATTGCCGCTATCCGCCCACAGTTCATAG |
| N88G-F | CTGGTGAAACAGGGTAGCAGCAATGTGGGCTATAGCTATG |
| N88G-R | GCCCACATTGCTGCTACCCTGTTTCACCAGTTTAAAATCCGC |
Table 1 Primers used by site-directed mutagenesis
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') |
|---|---|
| N37P-F | CGGATAGCGGCCCGAATAGCGCGACCTTTTATA-G |
| N37P-R | CGCGCTATTCGGGCCGCTATCCGCCCACAGTTCATAG |
| N38V-F | CGGATAGCGGCAATGTTAGCGCGACCTTTTATAGCGATGGC |
| N38V-R | GGTCGCGCTGTTATTGCCGCTATCCGCCCACAGTTCATAG |
| N88G-F | CTGGTGAAACAGGGTAGCAGCAATGTGGGCTATAGCTATG |
| N88G-R | GCCCACATTGCTGCTACCCTGTTTCACCAGTTTAAAATCCGC |

Fig. 1 Sequence analysis and screening of mutation sites A: Analysis of RMSF values of wild-type CDBFV at 298 K and 360 K. B: N-terminal sequence alignment of wild-type CDBFV with homologous xylanases, XynSW1, Xyn11B, Xyn11NX and PVX from Streptomyces sp., Aspergillus niger, Nesterenkonia xinjiangensis and Paecilonyces variotii, respectively. Green box refers to motif 36GNNS39. C: The flexible region 151SIDGD155和36GNNS39 with high RMSF value is highlighted in yellow in the structure
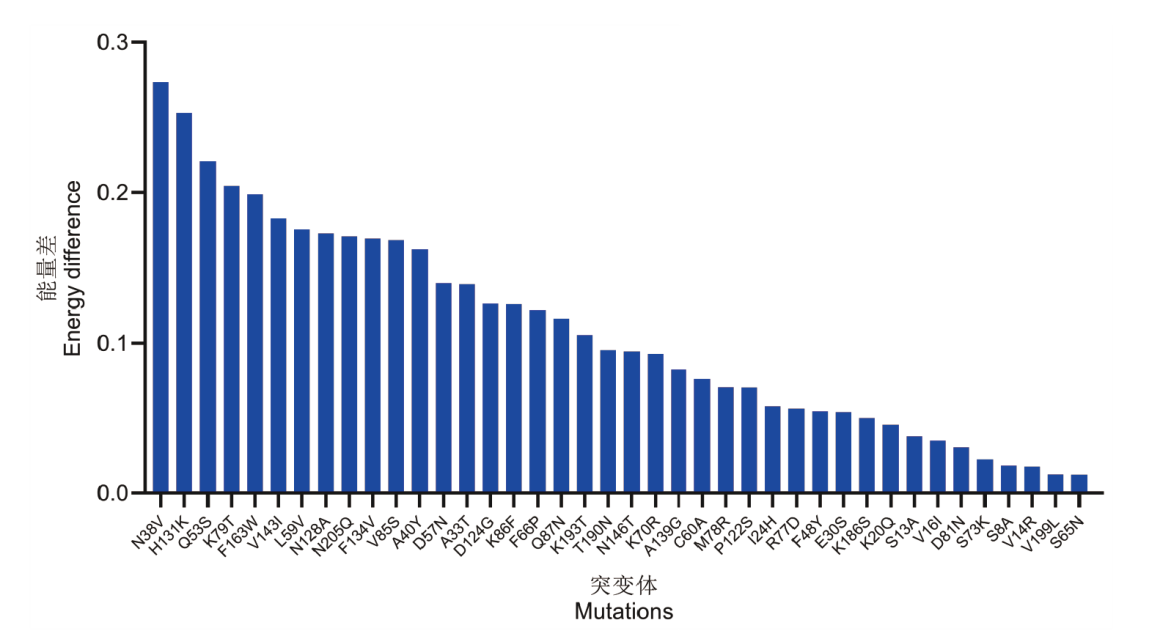
Fig. 2 Machine learning-based energy ranking prediction The figure illustrates the ranking of the most conserved amino acid scores at high temperature minus the wild-type amino acid scores predicted by a machine learning model
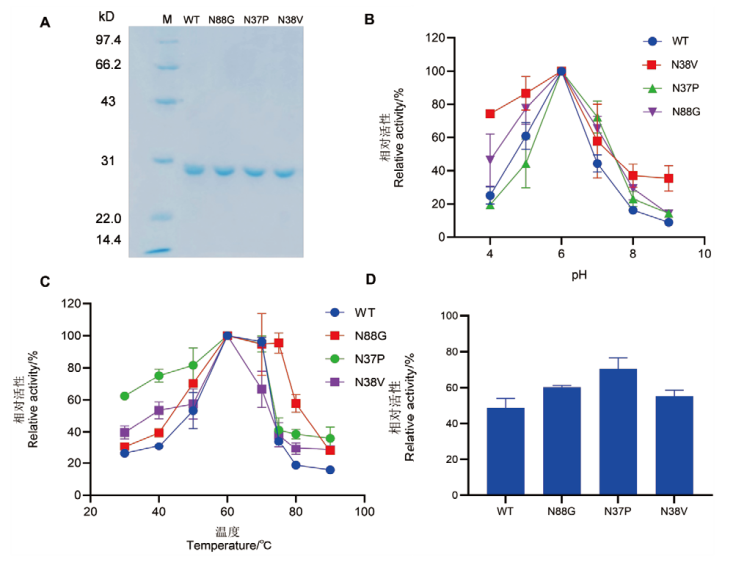
Fig. 3 SDS-PAGE analysis and enzymatic properties of wild-type and single mutants A: SDS-PAGE analysis of wild type CDBFV and single mutant; M: standard molecular weight protein marker. B: The optimal pH of wild type and single mutants. C: The optimal temperature of wild type and single mutants. D: The relative enzyme activity of wild type and single mutants after treatment at 85℃ for 3 min compared to their pre-treatment states
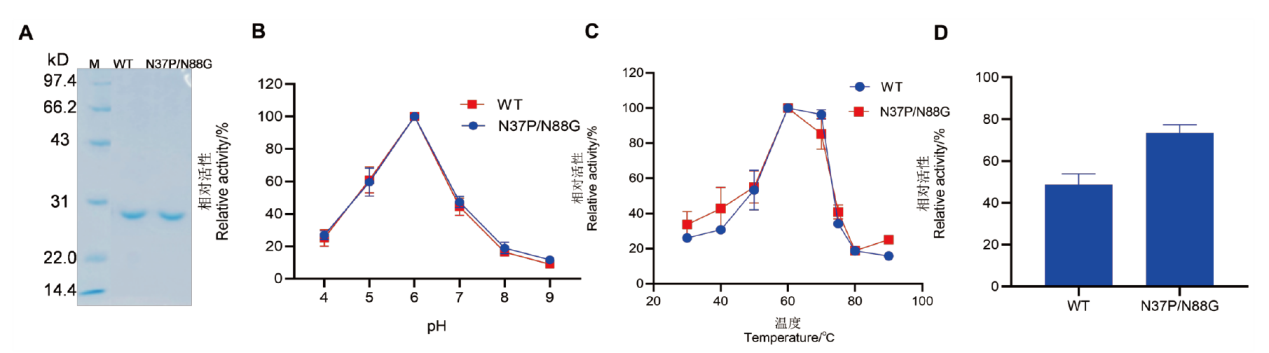
Fig. 4 SDS-PAGE analysis and enzymatic properties of wild-type and the double mutant A: SDS-PAGE analysis of wild type CDBFV and N37P/N88G double mutant; M: standard molecular weight protein marker. B: The optimal pH of wild type and the double mutant. C: The optimal temperature of wild type and the double mutant. D: The relative enzyme activity of wild type and the double mutant after treatment at 85℃ for 3 min compared to their pre-treatment states
| 酶Enzyme | 最适pH Optimal pH | 最适温度Optimal temperature/℃ | 绝对酶活Specific activity/(U·mg-1) | 相对酶活a Relative activity/% |
|---|---|---|---|---|
| WT(E. coli) | 6 | 60 | 5 216.2 | 48.7 |
| N37P(E. coli) | 6 | 60 | 1 108 | 70.3 |
| N38V(E. coli) | 6 | 60 | 2 130.45 | 55.1 |
| N37P/N88G(E. coli) | 6 | 60 | 807.2 | 73.4 |
| WT(P. pastoris) | 5 | 70 | 1 050 | 76.6 |
| N37P/N88G(P. pastoris) | 5 | 70 | 900 | 88.8 |
Table 2 Optimal condition and enzymatic activity determination of CDBFV wild-type and mutants
| 酶Enzyme | 最适pH Optimal pH | 最适温度Optimal temperature/℃ | 绝对酶活Specific activity/(U·mg-1) | 相对酶活a Relative activity/% |
|---|---|---|---|---|
| WT(E. coli) | 6 | 60 | 5 216.2 | 48.7 |
| N37P(E. coli) | 6 | 60 | 1 108 | 70.3 |
| N38V(E. coli) | 6 | 60 | 2 130.45 | 55.1 |
| N37P/N88G(E. coli) | 6 | 60 | 807.2 | 73.4 |
| WT(P. pastoris) | 5 | 70 | 1 050 | 76.6 |
| N37P/N88G(P. pastoris) | 5 | 70 | 900 | 88.8 |
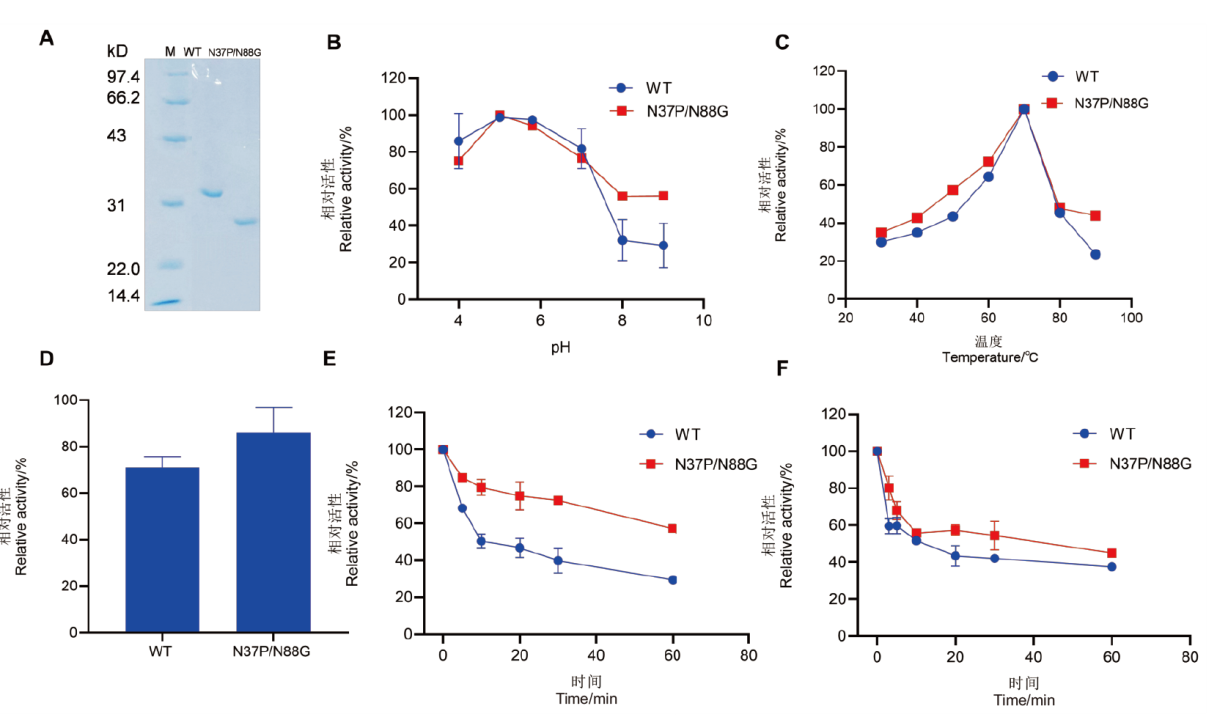
Fig. 5 SDS-PAGE analysis and enzymatic properties of wild-type and the double mutant expressed in Pichia pastoris A: SDS-PAGE analysis of wild type and double mutant; M: standard molecular weight protein marker. B: Optimal pH of wild type and the double mutant. C: Optimal temperature of wild type and the double mutant. D: The relative enzyme activity of wild type and the double mutant after treatment at 85℃ for 3 min compared to their pre-treatment states. E: Temperature stability of wild type and the double mutant after treatment at 70℃ for 0-60 min. F: Temperature stability of wild type and the double mutant after treatment at 80℃ for 0-60 min

Fig. 6 Structural analysis of the wild-type and double mutant N88G/N37P A: Analysis of the surface potential of the wild-type and mutant N88G mutant; blue, red and white colors correspond to positive, negative and uncharged regions, respectively. B: Amino acid interaction analysis of the wild-type and mutant N37P; the blue line indicates the hydrogen bond interactions
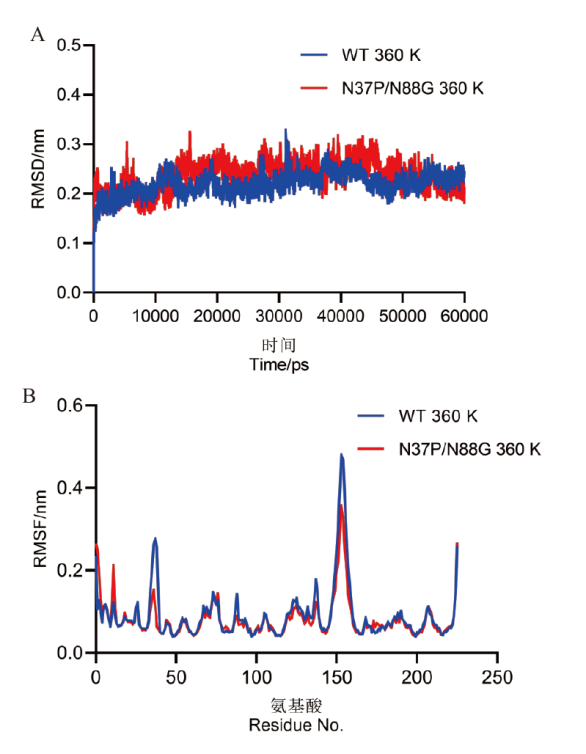
Fig. 7 Molecular dynamic simulation of the wild-type and double mutant at 360 K A: Analysis of the RMSD values for the wild-type and double mutant during a 50 ns simulation at 360 K. B: Analysis of the RMSF values for different residues in the wild-type and double mutant at 360 K
| [1] |
Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases[J]. FEMS Microbiol Rev, 2005, 29(1): 3-23.
doi: 10.1016/j.femsre.2004.06.005 pmid: 15652973 |
| [2] |
Subramaniyan S, Prema P. Biotechnology of microbial xylanases: enzymology, molecular biology, and application[J]. Crit Rev Biotechnol, 2002, 22(1): 33-64.
doi: 10.1080/07388550290789450 pmid: 11958335 |
| [3] | Mamo G. Alkaline active hemicellulases[J]. Adv Biochem Eng Biotechnol, 2020, 172: 245-291. |
| [4] |
Polizeli MLTM, Rizzatti ACS, Monti R, et al. Xylanases from fungi: properties and industrial applications[J]. Appl Microbiol Biotechnol, 2005, 67(5): 577-591.
doi: 10.1007/s00253-005-1904-7 pmid: 15944805 |
| [5] | Fernandes de Souza H, Aguiar Borges L, Dédalo Di Próspero Gonçalves V, et al. Recent advances in the application of xylanases in the food industry and production by Actinobacteria: a review[J]. Food Res Int, 2022, 162(Pt B): 112103. |
| [6] | Murthy PS, Naidu MM. Production and application of xylanase from Penicillium sp. utilizing coffee by-products[J]. Food Bioprocess Technol, 2012, 5(2): 657-664. |
| [7] | Motta FL, Andrade CCP, Sant MHA. A review of xylanase production by the fermentation of xylan: classification, characterization and applications[M]//Sustainable Degradation of Lignocellulosic Biomass - Techniques, Applications and Commercialization.: InTech, 2013. |
| [8] |
Woyengo TA, Sands JS, Guenter W, et al. Nutrient digestibility and performance responses of growing pigs fed phytase- and xylanase-supplemented wheat-based diets[J]. J Anim Sci, 2008, 86(4): 848-857.
doi: 10.2527/jas.2007-0018 pmid: 18203976 |
| [9] | Khandeparkar R, Bhosle NB. Application of thermoalkalophilic xylanase from Arthrobacter sp. MTCC 5214 in biobleaching of kraft pulp[J]. Bioresour Technol, 2007, 98(4): 897-903. |
| [10] | Liu SY, Zhuang XH, Zhang XL, et al. Enzymatic modification of rice bran polysaccharides by enzymes from Grifola frondosa: natural killer cell cytotoxicity and antioxidant activity[J]. J Food Sci, 2018, 83(7): 1948-1955. |
| [11] | Zhang Y, Guan FF, Xu GS, et al. A novel thermophilic chitinase directly mined from the marine metagenome using the deep learning tool Preoptem[J]. Bioresour Bioprocess, 2022, 9(1): 54. |
| [12] | Newberry RW, Raines RT. The n→π* interaction[J]. Acc Chem Res, 2017, 50(8): 1838-1846. |
| [13] | Han NY, Ma Y, Mu YL, et al. Enhancing thermal tolerance of a fungal GH11 xylanase guided by B-factor analysis and multiple sequence alignment[J]. Enzyme Microb Technol, 2019, 131: 109422. |
| [14] | Bajaj P, Mahajan R. Cellulase and xylanase synergism in industrial biotechnology[J]. Appl Microbiol Biotechnol, 2019, 103(21/22): 8711-8724. |
| [15] |
Nawab A, Ibtisham F, Li GH, et al. Heat stress in poultry production: mitigation strategies to overcome the future challenges facing the global poultry industry[J]. J Therm Biol, 2018, 78: 131-139.
doi: S0306-4565(18)30130-X pmid: 30509629 |
| [16] |
刘伟, 于凤民, 马千鹏, 等. 木聚糖酶在青贮及反刍动物日粮中的应用[J]. 草地学报, 2024, 32(1): 331-339.
doi: 10.11733/j.issn.1007-0435.2024.01.034 |
|
Liu W, Yu FM, Ma QP, et al. Application of xylanase in silage and ruminant diet[J]. Acta Agrestia sinica, 2024, 32(1): 331-339.
doi: 10.11733/j.issn.1007-0435.2024.01.034 |
|
| [17] | Alponti JS, Fonseca Maldonado R, Ward RJ. Thermostabilization of Bacillus subtilis GH11 xylanase by surface charge engineering[J]. Int J Biol Macromol, 2016, 87: 522-528. |
| [18] | Mendonça M, Barroca M, Collins T. Endo-1, 4-β-xylanase-containing glycoside hydrolase families: characteristics, singularities and similarities[J]. Biotechnol Adv, 2023, 65: 108148. |
| [19] | Chadha BS, Kaur B, Basotra N, et al. Thermostable xylanases from thermophilic fungi and bacteria: current perspective[J]. Bioresour Technol, 2019, 277: 195-203. |
| [20] | Zhang S, Zhang K, Chen XZ, et al. Five mutations in N-terminus confer thermostability on mesophilic xylanase[J]. Biochem Biophys Res Commun, 2010, 395(2): 200-206. |
| [21] | Wu H, Chen QM, Zhang WL, et al. Overview of strategies for developing high thermostability industrial enzymes: discovery, mechanism, modification and challenges[J]. Crit Rev Food Sci Nutr, 2023, 63(14): 2057-2073. |
| [22] | Li YY, Li C, Huang H, et al. Significantly enhanced thermostability of Aspergillus niger xylanase by modifying its highly flexible regions[J]. J Agric Food Chem, 2022, 70(15): 4620-4630. |
| [23] | You C, Huang Q, Xue HP, et al. Potential hydrophobic interaction between two cysteines in interior hydrophobic region improves thermostability of a family 11 xylanase from Neocallimastix patriciarum[J]. Biotechnol Bioeng, 2010, 105(5): 861-870. |
| [24] | Wu QH, Zhang CN, Dong WQ, et al. Simultaneously enhanced thermostability and catalytic activity of xylanase from Streptomyces rameus L2001 by rigidifying flexible regions in loop regions of the N-terminus[J]. J Agric Food Chem, 2023, 71(34): 12785-12796. |
| [25] | Bhat SK, Purushothaman K, Kini KR, et al. Design of mutants of GH11 xylanase from Bacillus pumilus for enhanced stability by amino acid substitutions in the N-terminal region: an in silico analysis[J]. J Biomol Struct Dyn, 2022, 40(17): 7666-7679. |
| [26] |
Foroozandeh Shahraki M, Farhadyar K, Kavousi K, et al. A generalized machine-learning aided method for targeted identification of industrial enzymes from metagenome: A xylanase temperature dependence case study[J]. Biotechnol Bioeng, 2021, 118(2): 759-769.
doi: 10.1002/bit.27608 pmid: 33095441 |
| [27] |
Eijsink VGH, Bjørk A, Gåseidnes S, et al. Rational engineering of enzyme stability[J]. J Biotechnol, 2004, 113(1-3): 105-120.
doi: 10.1016/j.jbiotec.2004.03.026 pmid: 15380651 |
| [28] |
Zhou XX, Wang YB, Pan YJ, et al. Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins[J]. Amino Acids, 2008, 34(1): 25-33.
pmid: 17710363 |
| [29] |
Karbalaei M, Rezaee SA, Farsiani H. Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins[J]. J Cell Physiol, 2020, 235(9): 5867-5881.
doi: 10.1002/jcp.29583 pmid: 32057111 |
| [30] | 朱泰承, 李寅. 毕赤酵母表达系统发展概况及趋势[J]. 生物工程学报, 2015, 31(6): 929-938. |
| Zhu TC, Li Y. Recent development of Pichia pastoris system: current status and future perspective[J]. Chin J Biotechnol, 2015, 31(6): 929-938. | |
| [31] |
Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs[J]. J Mol Biol, 1997, 269(4): 631-643.
doi: 10.1006/jmbi.1997.1042 pmid: 9217266 |
| [1] | CAI Yi-an, ZHANG Yi-qun, YANG Zi-xuan, LIU Ye-xue, LIU Wen-long, LU Fu-ping, LI Yu. Enhanced Expression of Protease K in Pichia pastoris through Molecular Chaperones and Analysis of Its Effect on Wool Scale Layer [J]. Biotechnology Bulletin, 2024, 40(7): 307-313. |
| [2] | JIANG Wen-ping, RAN Qiu-ping, LIU Jia-shu, ZHANG Hui-min, ZHANG Di, JIANG Zheng-bing, LI Hua-nan. Effects of Carbohydrate-binding Modules on the Enzymatic Properties of Xylanase [J]. Biotechnology Bulletin, 2024, 40(5): 269-279. |
| [3] | RUZHA Yelizhati, YANG Yu. Strategies for Increasing Heterologous Protein Expression in Pichia pastoris [J]. Biotechnology Bulletin, 2024, 40(3): 118-134. |
| [4] | XIA Guang-li, CAO Na, SUN Hui-hui, ZHAO Ling, CAO Rong. Advances in the Biological Modification of Galactose Oxidase [J]. Biotechnology Bulletin, 2024, 40(1): 176-185. |
| [5] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [6] | DONG Cong, GAO Qing-hua, WANG Yue, LUO Tong-yang, WANG Qing-qing. Increasing the Expression of FAD-dependent Glucose Dehydrogenase by Recombinant Pichia pastoris Using a Combined Strategy [J]. Biotechnology Bulletin, 2023, 39(6): 316-324. |
| [7] | QU Ge, SUN Zhou-tong. Catalytic Promiscuity-driven Redesign of Enzyme Functions [J]. Biotechnology Bulletin, 2023, 39(4): 1-9. |
| [8] | YANG Jun-zhao, ZHANG Xin-rui, SUN Qing-yang, ZHENG Fei. Affecting Mechanism of Loop B3 on the Function of GH7 Endoglucanase [J]. Biotechnology Bulletin, 2023, 39(10): 281-291. |
| [9] | WANG Yue, GAO Qing-hua, DONG Cong, LUO Tong-yang, WANG Qing-qing. Expression of Pyranose Oxidase with Optimized Codon in Pichia pastoris [J]. Biotechnology Bulletin, 2022, 38(4): 269-277. |
| [10] | LI Zhi-hao, ZHANG Ge, MO Zhi-jie, DENG Shuai-jun, LI Jia-yi, ZHANG Hai-bo, LIU Xiao-hui, LIU Hao-bao. Effects of a Xylanase-producing Bacillus cereus on the Composition and Fermented Products of Cigar Leaves [J]. Biotechnology Bulletin, 2022, 38(2): 105-112. |
| [11] | ZHANG Chen, ZHANG Tong-tong, LIU Hai-ping. Screening and Identification of Ethylene-forming Enzymes with High Activity and Thermostability [J]. Biotechnology Bulletin, 2022, 38(11): 269-276. |
| [12] | CHEN Chun, SU Ling-qia, XIA Wei, WU Jing. Improved the Thermostability of MTHase from Arthrobacter ramosus by Directed Evolution [J]. Biotechnology Bulletin, 2021, 37(3): 84-91. |
| [13] | YANG Yue, TAO Yan, XIE Jing, QIAN Yun-fang. Biosynthesis of Ctenopharyngodon idella C-type Lysozyme Based on Recombinant Pichia pastoris and Its Antibacterial Activity [J]. Biotechnology Bulletin, 2021, 37(12): 169-179. |
| [14] | WU Jiao, YU Gui-zhen, YUAN Hang, LIU Xian, GAO Yan-xiu, GONG Ming, ZOU Zhu-rong. Improvement on the Thermostability of Target Proteins by Fusing Rubredoxin from Hyperthermophile Pyrococcus furiosus [J]. Biotechnology Bulletin, 2021, 37(10): 110-119. |
| [15] | HUANG Kun-long, SU Xiao-yun, YAO Bin. Fusion Expression of Somatostatin with a Thermostable Xylanase and Characterization of the Fusion Protein [J]. Biotechnology Bulletin, 2020, 36(9): 235-243. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||