








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 173-183.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1576
Previous Articles Next Articles
GUO Ai1( ), JIANG Mu-yan1, Ha Li-ma-ti·Ba He-tai-li1, LIU Yu-yuan1, WANG Jing1,2(
), JIANG Mu-yan1, Ha Li-ma-ti·Ba He-tai-li1, LIU Yu-yuan1, WANG Jing1,2( )
)
Received:2021-12-21
Online:2022-10-26
Published:2022-11-11
Contact:
WANG Jing
E-mail:1061123080@qq.com;wangjing_imu@163.com
GUO Ai, JIANG Mu-yan, Ha Li-ma-ti·Ba He-tai-li, LIU Yu-yuan, WANG Jing. Transcriptome Analysis of Lycium ruthenicum Murr. Shoots in Anthocyanin Biosynthesis Response to Salt Stress[J]. Biotechnology Bulletin, 2022, 38(10): 173-183.
| 基因ID Gene ID | 序列Sequence(5'-3') |
|---|---|
| LrH2B1 | F:AGTGCTTCCTGGTGAATTGG |
| R:TGGATAATACCTAGCCCTAGTTTCC | |
| Cluster-45.236520 | F:TTTGCCTCTGGCTTTGTCTACC |
| R:TGGATACTGCTGTTGCCTTCAT | |
| Cluster-45.192144 | F:TGCACTGTTTGTCTCCTAGCCTCC |
| R:ACGCCATTCTTCTCGTCTCCATC | |
| Cluster-45.35768 | F:AAAACAGGACATGGCCTTTATG |
| R:CTGAGTTTGTAGGCAAGGTGGA | |
| Cluster-45.156739 | F:GCACAAAGTCTCCGAGAAGG |
| R:GGGGGCTTCATATTTCTTGTT | |
| Cluster-45.33457 | F:GAAGCCAACAGGTGAAACGACG |
| R:TCTTCATCAAAGCTCCCAAATGCT | |
| Cluster-45.168632 | F:GGTTTGATGAAAGGGGTGAAGAA |
| R:CCTGACGTGGCAGTATCGCTGAG, | |
| Cluster-45.207146 | F:TGAAGAATTAAGAGAGTCGAGGTCG |
| R:AAGTTGGCAATGGTGATGGTGTC. | |
| Cluster-45.136887 | F:TCAGATGAACCCAATTCACCAA |
| R:ACCCTAAAGAGCTGTCGGATGT | |
| Cluster-45.146553 | F:GGCACAGTAGCTGAATCCAAGA |
| R:CCTTTACCAACTTCTGCCCATA | |
| Cluster-45.150170 | F:GAGCAAGGCAGCAATAGCAGAT |
| R:AAGGTGGTGACATTGAGGTGAAG | |
| Cluster-45.146155 | F:GGTCAAGCCACTGATAGCCACA |
| R:GCATTCCTGCTTTCCCTGTGAT | |
| Cluster-45.117764 | F:TATCTTTCTTAGGCCCTTCCCTC |
| R:GCGTTATTGTTATTATTGTCGGTGT |
Table 1 List of specific primers for qPCR
| 基因ID Gene ID | 序列Sequence(5'-3') |
|---|---|
| LrH2B1 | F:AGTGCTTCCTGGTGAATTGG |
| R:TGGATAATACCTAGCCCTAGTTTCC | |
| Cluster-45.236520 | F:TTTGCCTCTGGCTTTGTCTACC |
| R:TGGATACTGCTGTTGCCTTCAT | |
| Cluster-45.192144 | F:TGCACTGTTTGTCTCCTAGCCTCC |
| R:ACGCCATTCTTCTCGTCTCCATC | |
| Cluster-45.35768 | F:AAAACAGGACATGGCCTTTATG |
| R:CTGAGTTTGTAGGCAAGGTGGA | |
| Cluster-45.156739 | F:GCACAAAGTCTCCGAGAAGG |
| R:GGGGGCTTCATATTTCTTGTT | |
| Cluster-45.33457 | F:GAAGCCAACAGGTGAAACGACG |
| R:TCTTCATCAAAGCTCCCAAATGCT | |
| Cluster-45.168632 | F:GGTTTGATGAAAGGGGTGAAGAA |
| R:CCTGACGTGGCAGTATCGCTGAG, | |
| Cluster-45.207146 | F:TGAAGAATTAAGAGAGTCGAGGTCG |
| R:AAGTTGGCAATGGTGATGGTGTC. | |
| Cluster-45.136887 | F:TCAGATGAACCCAATTCACCAA |
| R:ACCCTAAAGAGCTGTCGGATGT | |
| Cluster-45.146553 | F:GGCACAGTAGCTGAATCCAAGA |
| R:CCTTTACCAACTTCTGCCCATA | |
| Cluster-45.150170 | F:GAGCAAGGCAGCAATAGCAGAT |
| R:AAGGTGGTGACATTGAGGTGAAG | |
| Cluster-45.146155 | F:GGTCAAGCCACTGATAGCCACA |
| R:GCATTCCTGCTTTCCCTGTGAT | |
| Cluster-45.117764 | F:TATCTTTCTTAGGCCCTTCCCTC |
| R:GCGTTATTGTTATTATTGTCGGTGT |
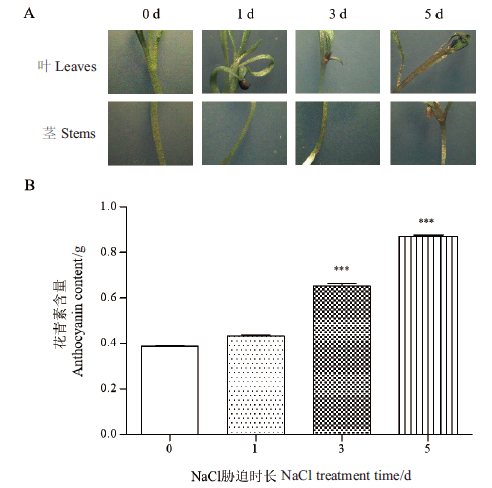
Fig. 1 Contents of anthocyanin in L. ruthenicum seedlings under NaCl stress at different times ***means the difference is extremely significant (P<0.001)
| 物质Compound | CK含量CK content/(μg·g-1) | Na含量Na content/(μg·g-1) | P值P -value | 倍数变化Fold change |
|---|---|---|---|---|
| 飞燕草素-3-O-葡萄糖苷 Delphinidin-3-O-glucoside | 0.151±0.0409 | 1.13±0.0283 | 0.000027 | 7.5 |
| 飞燕草素-3-O-芸香糖苷 Delphinidin-3-O-rutinoside | 0.432±0.0581 | 5.15±0.134 | 0.0000655 | 11.9 |
| 矮牵牛素-3-O-芸香糖苷 Petunidin-3-O-rutinoside | 2.33±0.323 | 5.313±0.286 | 0.000659 | 2.28 |
| 柚皮素-7-O-葡萄糖苷 Naringenin-7-O-glucoside | 0.112±0.0189 | 0.339±0.0457 | 0.0015 | 3.02 |
Table 2 Types and contents of significantly-changed anthocyanins in L. ruthenicum shoots under NaCl stress
| 物质Compound | CK含量CK content/(μg·g-1) | Na含量Na content/(μg·g-1) | P值P -value | 倍数变化Fold change |
|---|---|---|---|---|
| 飞燕草素-3-O-葡萄糖苷 Delphinidin-3-O-glucoside | 0.151±0.0409 | 1.13±0.0283 | 0.000027 | 7.5 |
| 飞燕草素-3-O-芸香糖苷 Delphinidin-3-O-rutinoside | 0.432±0.0581 | 5.15±0.134 | 0.0000655 | 11.9 |
| 矮牵牛素-3-O-芸香糖苷 Petunidin-3-O-rutinoside | 2.33±0.323 | 5.313±0.286 | 0.000659 | 2.28 |
| 柚皮素-7-O-葡萄糖苷 Naringenin-7-O-glucoside | 0.112±0.0189 | 0.339±0.0457 | 0.0015 | 3.02 |
| 样品Sample | 原始测序数据Raw reads | 过滤后测序数据Clean reads | 过滤后数据Clean bases | Error/% | Q20/% | Q30/% | GC content/% |
|---|---|---|---|---|---|---|---|
| CK1 | 55 697 476 | 54 900 826 | 8.24 G | 0.02 | 98.48 | 95.29 | 42.65 |
| CK2 | 60 551 930 | 59 757 658 | 8.96 G | 0.03 | 97.2 | 91.95 | 42.48 |
| CK3 | 55 743 892 | 54 933 528 | 8.24 G | 0.03 | 97.25 | 92.05 | 42.53 |
| Na1 | 61 583 804 | 61 024 066 | 9.15 G | 0.03 | 96.83 | 91.12 | 42.21 |
| Na2 | 46 367 542 | 45 803 662 | 6.87 G | 0.02 | 98.39 | 95.06 | 42.32 |
| Na3 | 47 635 904 | 46 871 318 | 7.03 G | 0.02 | 98.48 | 95.32 | 42.31 |
Table 3 Transcriptome sequencing data of L. ruthenicum shoots under control and NaCl stress
| 样品Sample | 原始测序数据Raw reads | 过滤后测序数据Clean reads | 过滤后数据Clean bases | Error/% | Q20/% | Q30/% | GC content/% |
|---|---|---|---|---|---|---|---|
| CK1 | 55 697 476 | 54 900 826 | 8.24 G | 0.02 | 98.48 | 95.29 | 42.65 |
| CK2 | 60 551 930 | 59 757 658 | 8.96 G | 0.03 | 97.2 | 91.95 | 42.48 |
| CK3 | 55 743 892 | 54 933 528 | 8.24 G | 0.03 | 97.25 | 92.05 | 42.53 |
| Na1 | 61 583 804 | 61 024 066 | 9.15 G | 0.03 | 96.83 | 91.12 | 42.21 |
| Na2 | 46 367 542 | 45 803 662 | 6.87 G | 0.02 | 98.39 | 95.06 | 42.32 |
| Na3 | 47 635 904 | 46 871 318 | 7.03 G | 0.02 | 98.48 | 95.32 | 42.31 |
| 分类Term | ID | P值P value | Q值Q value |
|---|---|---|---|
| 光合作用-天线蛋白Photosynthesis - antenna proteins | ko00196 | 1.38E-55 | 1.12E-53 |
| 光合作用Photosynthesis | ko00195 | 7.82E-14 | 3.17E-12 |
| 光合生物中碳的固定Carbon fixation in photosynthetic organisms | ko00710 | 3.80E-10 | 1.03E-08 |
| 植物激素信号转导Plant hormone signal transduction | ko04075 | 1.96E-08 | 3.96E-07 |
| 乙醛酸和二羧酸代谢Glyoxylate and dicarboxylate metabolism | ko00630 | 6.17E-07 | 9.99E-06 |
| 黄酮生物合成Flavonoid biosynthesis | ko00941 | 8.35E-07 | 1.13E-05 |
| 苯丙素生物合成Phenylpropanoid biosynthesis | ko00940 | 5.60E-06 | 6.48E-05 |
| 卟啉与叶绿素代谢Porphyrin and chlorophyll metabolism | ko00860 | 0.00041 | 0.0041 |
| 角质,木栓质和蜡质的生物合成Cutin,suberine and wax biosynthesis | ko00073 | 0.00045 | 0.0041 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | ko00500 | 0.001 | 0.0081 |
| 二苯乙烯类、二芳基庚烷类和姜辣素生物合成Stilbenoid,diarylheptanoid and gingerol biosynthesis | ko00945 | 0.0011 | 0.0081 |
| 氮代谢Nitrogen metabolism | ko00910 | 0.0046 | 0.031 |
| α-亚麻酸代谢alpha-linolenic acid metabolism | ko00592 | 0.005 | 0.031 |
| 精氨酸和脯氨酸代谢Arginine and proline metabolism | ko00330 | 0.0073 | 0.042 |
Table 4 KEGG enrichment pathways
| 分类Term | ID | P值P value | Q值Q value |
|---|---|---|---|
| 光合作用-天线蛋白Photosynthesis - antenna proteins | ko00196 | 1.38E-55 | 1.12E-53 |
| 光合作用Photosynthesis | ko00195 | 7.82E-14 | 3.17E-12 |
| 光合生物中碳的固定Carbon fixation in photosynthetic organisms | ko00710 | 3.80E-10 | 1.03E-08 |
| 植物激素信号转导Plant hormone signal transduction | ko04075 | 1.96E-08 | 3.96E-07 |
| 乙醛酸和二羧酸代谢Glyoxylate and dicarboxylate metabolism | ko00630 | 6.17E-07 | 9.99E-06 |
| 黄酮生物合成Flavonoid biosynthesis | ko00941 | 8.35E-07 | 1.13E-05 |
| 苯丙素生物合成Phenylpropanoid biosynthesis | ko00940 | 5.60E-06 | 6.48E-05 |
| 卟啉与叶绿素代谢Porphyrin and chlorophyll metabolism | ko00860 | 0.00041 | 0.0041 |
| 角质,木栓质和蜡质的生物合成Cutin,suberine and wax biosynthesis | ko00073 | 0.00045 | 0.0041 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | ko00500 | 0.001 | 0.0081 |
| 二苯乙烯类、二芳基庚烷类和姜辣素生物合成Stilbenoid,diarylheptanoid and gingerol biosynthesis | ko00945 | 0.0011 | 0.0081 |
| 氮代谢Nitrogen metabolism | ko00910 | 0.0046 | 0.031 |
| α-亚麻酸代谢alpha-linolenic acid metabolism | ko00592 | 0.005 | 0.031 |
| 精氨酸和脯氨酸代谢Arginine and proline metabolism | ko00330 | 0.0073 | 0.042 |
| 基因ID Gene ID | CK-FPKM | Na-FPKM | UP/DOWN | 倍数变化Fold change | Q值Q value | 参与激素通路Involved in hormonal pathway |
|---|---|---|---|---|---|---|
| Cluster-45.33300 | 2.8 | 67.88 | UP | 4.58 | 1.10E-43 | IAA |
| Cluster-45.160336 | 17.22 | 48.62 | UP | 1.48 | 2.68E-17 | Zeatin |
| Cluster-45.160335 | 4.75 | 12.57 | UP | 1.39 | 1.89E-12 | |
| Cluster-45.155979 | 1.25 | 18.36 | UP | 3.87 | 7.67E-37 | Gibberellin |
| Cluster-45.155981 | 2.33 | 28.19 | UP | 3.58 | 7.85E-39 | |
| Cluster-45.156292 | 43.04 | 18.56 | DOWN | -1.23 | 1.44E-10 | |
| Cluster-45.142941 | 19.73 | 1.94 | DOWN | -3.37 | 4.36E-25 | ABA |
| Cluster-45.123460 | 11.69 | 142.1 | UP | 3.59 | 2.95E-50 | |
| Cluster-45.195827 | 0.98 | 33.77 | UP | 5.07 | 1.29E-33 | |
| Cluster-45.39863 | 2.83 | 27.75 | UP | 3.29 | 1.69E-40 | |
| Cluster-45.78587 | 9.97 | 47.67 | UP | 2.24 | 8.10E-32 | |
| Cluster-45.80560 | 9.79 | 49.99 | UP | 2.34 | 9.08E-41 | |
| Cluster-45.80561 | 13.05 | 56.32 | UP | 2.09 | 2.47E-34 | |
| Cluster-45.165767 | 68.97 | 867.3 | UP | 3.58 | 3.94E-33 | |
| Cluster-45.165775 | 9.24 | 120.92 | UP | 3.68 | 2.67E-23 | |
| Cluster-45.165770 | 39.57 | 491.37 | UP | 3.6 | 1.60E-59 | |
| Cluster-45.165774 | 127.23 | 1730.95 | UP | 3.6 | 2.53E-05 | |
| Cluster-45.191345 | 1.03 | 31.02 | UP | 4.89 | 3.36E-12 | |
| Cluster-45.191350 | 1.29 | 13.64 | UP | 3.4 | 1.09E-09 | |
| Cluster-45.191353 | 17.41 | 102.01 | UP | 2.54 | 1.37E-49 | |
| Cluster-45.191354 | 2.84 | 17.66 | UP | 2.63 | 1.72E-09 | |
| Cluster-45.161351 | 96.31 | 244.8 | UP | 1.33 | 2.71E-19 | Ethylene |
| Cluster-45.161685 | 11.82 | 45.31 | UP | 1.91 | 1.87E-15 | |
| Cluster-45.108200 | 45.25 | 1.29 | DOWN | -5.16 | 8.45E-13 | |
| Cluster-45.32854 | 1.48 | 27.85 | UP | 4.21 | 2.26E-52 | Brassinosteroid |
| Cluster-45.175682 | 17.06 | 47.88 | UP | 1.47 | 1.34E-14 | SA |
| Cluster-45.181144 | 159.8 | 12.52 | DOWN | -3.7 | 1.37E-25 |
Table 5 DEGs related to hormone signaling transduction pathway in L. ruthenicum shoots under NaCl stress
| 基因ID Gene ID | CK-FPKM | Na-FPKM | UP/DOWN | 倍数变化Fold change | Q值Q value | 参与激素通路Involved in hormonal pathway |
|---|---|---|---|---|---|---|
| Cluster-45.33300 | 2.8 | 67.88 | UP | 4.58 | 1.10E-43 | IAA |
| Cluster-45.160336 | 17.22 | 48.62 | UP | 1.48 | 2.68E-17 | Zeatin |
| Cluster-45.160335 | 4.75 | 12.57 | UP | 1.39 | 1.89E-12 | |
| Cluster-45.155979 | 1.25 | 18.36 | UP | 3.87 | 7.67E-37 | Gibberellin |
| Cluster-45.155981 | 2.33 | 28.19 | UP | 3.58 | 7.85E-39 | |
| Cluster-45.156292 | 43.04 | 18.56 | DOWN | -1.23 | 1.44E-10 | |
| Cluster-45.142941 | 19.73 | 1.94 | DOWN | -3.37 | 4.36E-25 | ABA |
| Cluster-45.123460 | 11.69 | 142.1 | UP | 3.59 | 2.95E-50 | |
| Cluster-45.195827 | 0.98 | 33.77 | UP | 5.07 | 1.29E-33 | |
| Cluster-45.39863 | 2.83 | 27.75 | UP | 3.29 | 1.69E-40 | |
| Cluster-45.78587 | 9.97 | 47.67 | UP | 2.24 | 8.10E-32 | |
| Cluster-45.80560 | 9.79 | 49.99 | UP | 2.34 | 9.08E-41 | |
| Cluster-45.80561 | 13.05 | 56.32 | UP | 2.09 | 2.47E-34 | |
| Cluster-45.165767 | 68.97 | 867.3 | UP | 3.58 | 3.94E-33 | |
| Cluster-45.165775 | 9.24 | 120.92 | UP | 3.68 | 2.67E-23 | |
| Cluster-45.165770 | 39.57 | 491.37 | UP | 3.6 | 1.60E-59 | |
| Cluster-45.165774 | 127.23 | 1730.95 | UP | 3.6 | 2.53E-05 | |
| Cluster-45.191345 | 1.03 | 31.02 | UP | 4.89 | 3.36E-12 | |
| Cluster-45.191350 | 1.29 | 13.64 | UP | 3.4 | 1.09E-09 | |
| Cluster-45.191353 | 17.41 | 102.01 | UP | 2.54 | 1.37E-49 | |
| Cluster-45.191354 | 2.84 | 17.66 | UP | 2.63 | 1.72E-09 | |
| Cluster-45.161351 | 96.31 | 244.8 | UP | 1.33 | 2.71E-19 | Ethylene |
| Cluster-45.161685 | 11.82 | 45.31 | UP | 1.91 | 1.87E-15 | |
| Cluster-45.108200 | 45.25 | 1.29 | DOWN | -5.16 | 8.45E-13 | |
| Cluster-45.32854 | 1.48 | 27.85 | UP | 4.21 | 2.26E-52 | Brassinosteroid |
| Cluster-45.175682 | 17.06 | 47.88 | UP | 1.47 | 1.34E-14 | SA |
| Cluster-45.181144 | 159.8 | 12.52 | DOWN | -3.7 | 1.37E-25 |
| 基因ID Gene ID | CK-FPKM | Na-FPKM | UP/DOWN | 倍数变化 Fold change | Q值Q value | 转录因子类型Transcription factor type |
|---|---|---|---|---|---|---|
| Cluster-45.111653 | 4.48 | 17.95 | UP | 1.98 | 4.23E-17 | MYB transcripiton factor |
| Cluster-45.111657 | 6.51 | 27.56 | UP | 2.06 | 7.91E-17 | |
| Cluster-45.111658 | 10.05 | 33.49 | UP | 1.72 | 8.62E-21 | |
| Cluster-45.112186 | 18.55 | 56.83 | UP | 1.6 | 7.91E-16 | |
| Cluster-45.227246 | 5.38 | 38.31 | UP | 2.81 | 3.07E-32 | |
| Cluster-45.227247 | 1.4 | 8.37 | UP | 2.56 | 4.74E-20 | |
| Cluster-45.32932 | 7.36 | 30.01 | UP | 2.01 | 4.9E-14 | |
| Cluster-45.33457 | 4.23 | 43.42 | UP | 3.33 | 5.13E-34 | |
| Cluster-45.35768 | 2.26 | 61.07 | UP | 4.73 | 5.35E-32 | |
| Cluster-45.79986 | 4.58 | 19.6 | UP | 2.08 | 1.43E-15 | |
| Cluster-45.95693 | 79.87 | 270.84 | UP | 1.73 | 7.24E-23 | |
| Cluster-45.146155 | 40.64 | 7.09 | DOWN | -2.54 | 5.16E-20 | bHLH transcription factor |
| Cluster-45.147909 | 52.14 | 21.72 | DOWN | -1.28 | 1.09E-11 | |
| Cluster-45.150170 | 51.34 | 7.13 | DOWN | -2.87 | 9.35E-26 | |
| Cluster-45.161489 | 30.39 | 2.7 | DOWN | -3.52 | 1.76E-11 | |
| Cluster-45.168632 | 4.11 | 13.93 | UP | 1.75 | 4.3E-15 |
Table 6 MYB and bHLH transcription factors with significantly changed expression levels
| 基因ID Gene ID | CK-FPKM | Na-FPKM | UP/DOWN | 倍数变化 Fold change | Q值Q value | 转录因子类型Transcription factor type |
|---|---|---|---|---|---|---|
| Cluster-45.111653 | 4.48 | 17.95 | UP | 1.98 | 4.23E-17 | MYB transcripiton factor |
| Cluster-45.111657 | 6.51 | 27.56 | UP | 2.06 | 7.91E-17 | |
| Cluster-45.111658 | 10.05 | 33.49 | UP | 1.72 | 8.62E-21 | |
| Cluster-45.112186 | 18.55 | 56.83 | UP | 1.6 | 7.91E-16 | |
| Cluster-45.227246 | 5.38 | 38.31 | UP | 2.81 | 3.07E-32 | |
| Cluster-45.227247 | 1.4 | 8.37 | UP | 2.56 | 4.74E-20 | |
| Cluster-45.32932 | 7.36 | 30.01 | UP | 2.01 | 4.9E-14 | |
| Cluster-45.33457 | 4.23 | 43.42 | UP | 3.33 | 5.13E-34 | |
| Cluster-45.35768 | 2.26 | 61.07 | UP | 4.73 | 5.35E-32 | |
| Cluster-45.79986 | 4.58 | 19.6 | UP | 2.08 | 1.43E-15 | |
| Cluster-45.95693 | 79.87 | 270.84 | UP | 1.73 | 7.24E-23 | |
| Cluster-45.146155 | 40.64 | 7.09 | DOWN | -2.54 | 5.16E-20 | bHLH transcription factor |
| Cluster-45.147909 | 52.14 | 21.72 | DOWN | -1.28 | 1.09E-11 | |
| Cluster-45.150170 | 51.34 | 7.13 | DOWN | -2.87 | 9.35E-26 | |
| Cluster-45.161489 | 30.39 | 2.7 | DOWN | -3.52 | 1.76E-11 | |
| Cluster-45.168632 | 4.11 | 13.93 | UP | 1.75 | 4.3E-15 |
| [1] |
Liu JY, Osbourn A, Ma PD. MYB transcription factors as regulators of phenylpropanoid metabolism in plants[J]. Mol Plant, 2015, 8(5):689-708.
doi: 10.1016/j.molp.2015.03.012 pmid: 25840349 |
| [2] |
Griesbach RJ. Correlation of pH and light intensity on flower color in potted Eustoma grandiflorum grise[J]. HortScience, 1992, 27(7):817-818.
doi: 10.21273/HORTSCI.27.7.817 URL |
| [3] |
Li P, Li YJ, Zhang FJ, et al. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation[J]. Plant J, 2017, 89(1):85-103.
doi: 10.1111/tpj.13324 URL |
| [4] |
Perea-Resa C, Rodríguez-Milla MA, Iniesto E, et al. Prefoldins negatively regulate cold acclimation in Arabidopsis thaliana by promoting nuclear proteasome-mediated HY5 degradation[J]. Mol Plant, 2017, 10(6):791-804.
doi: S1674-2052(17)30101-6 pmid: 28412546 |
| [5] |
Su NN, Wu Q, Cui J. Increased sucrose in the hypocotyls of radish sprouts contributes to nitrogen deficiency-induced anthocyanin accumulation[J]. Front Plant Sci, 2016, 7:1976.
doi: 10.3389/fpls.2016.01976 pmid: 28083009 |
| [6] |
Neill SO, Gould KS. Anthocyanins in leaves:light attenuators or antioxidants?[J]. Funct Plant Biol, 2003, 30(8):865-873.
doi: 10.1071/FP03118 URL |
| [7] | Chalker-Scott L. Do anthocyanins function as osmoregulators in leaf tissues?[M]//Advances in Botanical Research. Amsterdam:Elsevier, 2002:103-127. |
| [8] |
McKown R, Kuroki G, Warren G. Cold responses of Arabidopsis mutants impaired in freezing tolerance[J]. J Exp Bot, 1996, 47(12):1919-1925.
doi: 10.1093/jxb/47.12.1919 URL |
| [9] |
Neill SO, Gould KS, Kilmartin PA, et al. Antioxidant activities of red versus green leaves in Elatostema rugosum[J]. Plant Cell Environ, 2002, 25(4):539-547.
doi: 10.1046/j.1365-3040.2002.00837.x URL |
| [10] |
Nagata T, Todoriki S, Masumizu T, et al. Levels of active oxygen species are controlled by ascorbic acid and anthocyanin in Arabidopsis[J]. J Agric Food Chem, 2003, 51(10):2992-2999.
doi: 10.1021/jf026179+ URL |
| [11] |
Li J, Zhao AC, Yu MD, et al. Function analysis of anthocyanidin synthase from Morus alba L. by expression in bacteria and tobacco[J]. Electron J Biotechnol, 2018, 36:9-14.
doi: 10.1016/j.ejbt.2018.09.001 URL |
| [12] |
Li JJ, Ma JJ, Guo HL, et al. Growth and physiological responses of two phenotypically distinct accessions of centipedegrass(Eremochloa ophiuroides(Munro)Hack. )to salt stress[J]. Plant Physiol Biochem, 2018, 126:1-10.
doi: 10.1016/j.plaphy.2018.02.018 URL |
| [13] | 张鹏, 贺虹, 聂福彪, 等. 民勤荒漠区黑果枸杞所在群落种间联结性研究[J]. 林业科技通讯, 2019(8):3-7. |
| Zhang P, He H, Nie FB, et al. Study on interspecific association of Lycium ruthenicum community in Minqin desert area[J]. For Sci Technol, 2019(8):3-7. | |
| [14] | 李善家. 黑河下游荒漠植物黑果枸杞分布格局及盐碱适应性研究[D]. 兰州: 兰州理工大学, 2017. |
| Li SJ. Study on the distribution pattern and salt-alkali adaptability of the desert plant Lycium ruthenicum in the lower reaches of the Heihe River[D]. Lanzhou: Lanzhou University of Technology, 2017. | |
| [15] | 姜霞, 任红旭, 马占青, 等. 黑果枸杞耐盐机理的相关研究[J]. 北方园艺, 2012(10):19-23. |
| Jiang X, Ren HX, Ma ZQ, et al. Studies on the physiological mechanism underlying salt tolerance of Lycium ruthenicum Murr[J]. North Hortic, 2012(10):19-23. | |
| [16] | 杨万鹏, 马瑞, 杨永义, 等. 不同浓度NaCl处理对黑果枸杞叶片性状的影响[J]. 生态科学, 2019, 38(4):35-41. |
| Yang WP, Ma R, Yang YY, et al. Effects of NaCl treatment on leaf traits of Lycium ruthenicum[J]. Ecol Sci, 2019, 38(4):35-41. | |
| [17] | 李岩, 何学敏, 杨晓东, 等. 不同生境黑果枸杞根际与非根际土壤微生物群落多样性[J]. 生态学报, 2018, 38(17):5983-5995. |
| Li Y, He XM, Yang XD, et al. The microbial community diversity of the rhizosphere and bulk soils of Lycium ruthenicum in different habitats[J]. Acta Ecol Sin, 2018, 38(17):5983-5995. | |
| [18] | 王龙强, 米永伟, 蔺海明. 盐胁迫对枸杞属两种植物幼苗离子吸收和分配的影响[J]. 草业学报, 2011, 20(4):129-136. |
| Wang LQ, Mi YW, Lin HM. Effect of salt stress on ion absorption and distribution of two Lycium seedlings[J]. Acta Prataculturae Sin, 2011, 20(4):129-136. | |
| [19] | 王静, 马腾斋, 邱佳俊, 等. NaCl胁迫对黑果枸杞幼苗生理及生化指标的影响[J]. 北方园艺, 2019(6):59-64. |
| Wang J, Ma TZ, Qiu JJ, et al. Effects of NaCl stress on physiological and biochemical characteristics of Lycium ruthecium Murr. seedling[J]. North Hortic, 2019(6):59-64. | |
| [20] | Oh JE. Enhanced level of anthocyanin leads to increased salt tolerance in Arabidopsis PAP1-D plants upon sucrose treatment[J]. J Korean Soc Appl Biol Chem, 2011, 54(1):79-88. |
| [21] |
Naing AH, Park KI, Ai TN, et al. Overexpression of snapdragon Delila(Del)gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance[J]. BMC Plant Biol, 2017, 17(1):65.
doi: 10.1186/s12870-017-1015-5 pmid: 28335727 |
| [22] | 孙红, 孙田雨, 许丽丽, 等. 长期低盐处理对葡萄果实品质及转录组的影响[J]. 植物生理学报, 2017, 53(12):2197-2205. |
| Sun H, Sun TY, Xu LL, et al. Effects of the long-term treatment of low-concentrated salt on grape berry quality and transcriptome[J]. Plant Physiol J, 2017, 53(12):2197-2205. | |
| [23] | 吕东林, 林琳, 郭译文, 等. 紫雨桦耐盐性及花色苷合成相关基因的表达特性[J]. 南京林业大学学报:自然科学版, 2018, 42(2):25-32. |
| Lyu DL, Lin L, Guo YW, et al. Characterization of gene expression in anthocyanin synthesis and salt tolerance of Betula pendula ‘Purple Rain’[J]. J Nanjing For Univ Nat Sci Ed, 2018, 42(2):25-32. | |
| [24] |
Zeng SH, Wu M, Zou CY, et al. Comparative analysis of anthocyanin biosynthesis during fruit development in two Lycium species[J]. Physiol Plant, 2014, 150(4):505-516.
doi: 10.1111/ppl.12131 URL |
| [25] |
Zhang YQ, Liu ZJ, Liu JP, et al. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation[J]. Plant Cell Rep, 2017, 36(4):557-569.
doi: 10.1007/s00299-017-2102-7 pmid: 28275852 |
| [26] |
Sun X, Jia X, Huo LQ, et al. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple[J]. Plant Cell Environ, 2018, 41(2):469-480.
doi: 10.1111/pce.13110 URL |
| [27] |
Xie XB, Li S, Zhang RF, et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples[J]. Plant Cell Environ, 2012, 35(11):1884-1897.
doi: 10.1111/j.1365-3040.2012.02523.x URL |
| [28] |
Hu DG, Yu JQ, Han PL, et al. The regulatory module MdPUB29-MdbHLH3 connects ethylene biosynthesis with fruit quality in apple[J]. New Phytol, 2019, 221(4):1966-1982.
doi: 10.1111/nph.15511 URL |
| [29] | 余建, 赵爱春, 刘长英, 等. 外源乙烯利与1-MCP处理对桑椹中乙烯和花青素相关代谢基因表达的影响[J]. 林业科学, 2017, 53(2):138-148. |
| Yu J, Zhao AC, Liu CY, et al. Effects of exogenous ethylene and 1-MCP treatments on the expression of genes involved in ethylene and anthocyanin in mulberry fruit[J]. Sci Silvae Sin, 2017, 53(2):138-148. | |
| [30] |
Song JY, Kim TY, Hong JH. Effects of abscisic acid and temperature on the anthocyanin accumulation in seedlings of Arabidopsis thaliana[J]. J Environ Sci Int, 2005, 14(12):1093-1102.
doi: 10.5322/JES.2005.14.12.1093 URL |
| [31] |
Oh HD, Yu DJ, Chung SW, et al. Abscisic acid stimulates anthocyanin accumulation in ‘Jersey’ highbush blueberry fruits during ripening[J]. Food Chem, 2018, 244:403-407.
doi: 10.1016/j.foodchem.2017.10.051 URL |
| [32] | 于淼, 刘海峰, 王军. ABA对葡萄花色苷合成相关基因表达的影响[J]. 果树学报, 2012, 29(1):29-35, 157. |
| Yu M, Liu HF, Wang J. Effects of ABA on expression of genes related to anthocyanin biosynthesis in grapevine[J]. J Fruit Sci, 2012, 29(1):29-35, 157. |
| [1] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [2] | SUN Wei, ZHANG Yan, WANG Yu-han, XU Hui, XU Xiao-rong, JU Zhi-gang. Cloning of Rd3GT1 in Rhododendron delavayi and Its Effect on Flower Color Formation of Petunia hybrida [J]. Biotechnology Bulletin, 2022, 38(9): 198-206. |
| [3] | DUAN Yue-tong, WANG Peng-nian, ZHANG Chun-bao, LIN Chun-jing. Research Progress in Plant Flavanone-3-hydroxylase Gene [J]. Biotechnology Bulletin, 2022, 38(6): 27-33. |
| [4] | ZOU Liang-ping, GUO Xin, QI Deng-feng, ZHAI Min, LI Zhuang, ZHAO Ping-juan, PENG Ming, NIU Xing-kui. Anthocyanin Accumulation and Its Gene Expression Induced by Low Nitrogen Stress in Cassava Seedlings [J]. Biotechnology Bulletin, 2022, 38(2): 75-82. |
| [5] | LUO Ya-fang, ZHU Chun-hua, XIAO Yu-ting, LI Fang-quan, ZHANG Jiang, WANG Yu-shu. Screening and Functional Analysis of UGT Genes Involved in the Flavonoid Biosynthesis of Brassica oleracea var. acephala [J]. Biotechnology Bulletin, 2022, 38(11): 194-201. |
| [6] | LU Wen-ying, ZHAO Lei, LI Tian-qi, CUI He-yun, LIAO Ping-an. Research Advances of Fruit Anthocyanin Accumulation in Rosaceae Plants [J]. Biotechnology Bulletin, 2021, 37(1): 234-245. |
| [7] | SU Jie, GUO Rong-qi, GAO Yang, YU Xiu-min, LI Guo-jing, WANG Rui-gang. Response to NaCl and ABA in Arabidopsis thaliana of the Double Silent Gene VHA-c2&c4 [J]. Biotechnology Bulletin, 2020, 36(7): 48-54. |
| [8] | GAO Guo-ying, WU Xiao-fang, ZHANG Da-wei, ZHOU Ding-gang, ZHANG Kai-xuan, YAN Ming-li. Research Progress on the MBW Complexes in Plant Anthocyanin Biosynthesis Pathway [J]. Biotechnology Bulletin, 2020, 36(1): 126-134. |
| [9] | ZHANG Meng-ying, LI Ya-hui, ZHAN Yuan-long, LIU Chang-li. Research Advances in Biosynthesis of Polyhydroxyalkanoates(PHAs)by Halophilic Bacteria [J]. Biotechnology Bulletin, 2019, 35(6): 172-177. |
| [10] | LI Chong-hui, YIN Jun-mei. Genetic Engineering Progress and Breeding Tactics on Blue Flowers [J]. Biotechnology Bulletin, 2019, 35(11): 160-168. |
| [11] | ZHAO Xiang-jie, YANG Wen-jun, YANG Rong-ling, WU Ting-ting, WANG Zhao-yu, XU Ning-ning, HE Jia-mei. Research Progress on Biotransformation Modification of Anthocyanins [J]. Biotechnology Bulletin, 2019, 35(10): 205-211. |
| [12] | LIU Xiao-wei, YANG Xiu-yan, WU Hai-wen, ZHI Xiao-rong, ZHU Jian-feng, ZHANG Hua-xin. Effects of NaCl Stress on the Germination of Reaumuria soongorica and Evaluation of Salt Tolerance at Germination Stage [J]. Biotechnology Bulletin, 2019, 35(1): 27-34. |
| [13] | WANG Wei-guo, DENG Qian, JI Qiao-ling, WANG Wei. Orthogonal Test of EMS Mutation and NaCl Stress and Screening of Salt-tolerant Fiber Flax Germplasm in Vitro [J]. Biotechnology Bulletin, 2016, 32(6): 103-110. |
| [14] | Jin Qifang, Sun Weijiang, Chen Zhidan. Regulation Mechanism of Anthocyanin Synthesis in Purple Shoots of Tea by Lighting [J]. Biotechnology Bulletin, 2015, 31(6): 20-27. |
| [15] | Xiao Xiangwen, Zhu Qilang, Liu Haifeng, Wang Junduo, Luo Cheng, Zeng Wen, Liang Yajun, Gong Zhaolong, Li Xiaobo. Cloning and Bioinformatics Analysis of DFR Gene from Brown Cotton(Gossypium hirsutum L.)Fiber [J]. Biotechnology Bulletin, 2014, 0(5): 88-95. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||