








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (12): 20-33.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0392
Previous Articles Next Articles
ZHOU Zi-ying1( ), SONG Xiao-dong2, LIU Yang-er3,4, WU Yi-fan3, ZHU Long-jiao3, GU Dong-yue5, HE Guo-qing6, LI Xiang-yang1(
), SONG Xiao-dong2, LIU Yang-er3,4, WU Yi-fan3, ZHU Long-jiao3, GU Dong-yue5, HE Guo-qing6, LI Xiang-yang1( ), XU Wen-tao3(
), XU Wen-tao3( )
)
Received:2024-04-26
Online:2024-12-26
Published:2025-01-15
Contact:
LI Xiang-yang, XU Wen-tao
E-mail:18210727086@163.com;lxy2002cn@163.com;xuwentao@cau.edu.cn
ZHOU Zi-ying, SONG Xiao-dong, LIU Yang-er, WU Yi-fan, ZHU Long-jiao, GU Dong-yue, HE Guo-qing, LI Xiang-yang, XU Wen-tao. Construction Strategies of Allosteric Transcription Factor Biosensors and Their Application Advances in Food Safety[J]. Biotechnology Bulletin, 2024, 40(12): 20-33.
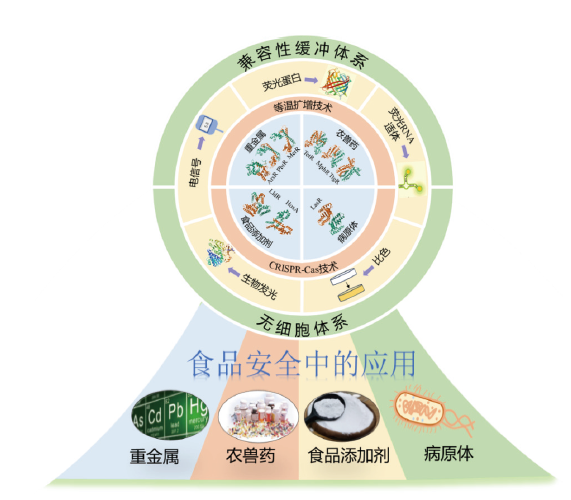
Fig. 1 Construction strategy of aTF biosensor and its application in food safety A typical system of aTF biosensor includes a molecular recognition element-aTF, a signal amplification strategy, a signal output system and a sensing system. The constructed aTF biosensor can be applied to detect heavy metals, pesticide and veterinary drug residues, food additives and pathogens in food and other food safety fields
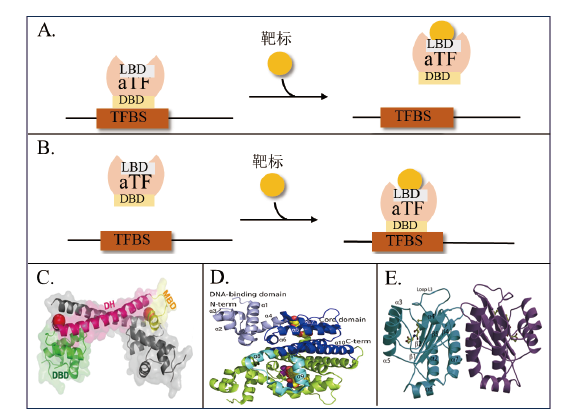
Fig. 2 Schematic diagram of the mechanism of metamorphic regulation of aTF and structure of aTF A: Repressed aTF regulatory mechanism. B: Activated aTF regulatory mechanism. C: Stereoview of the overall structure of mercury(II)-bound Tn501 MerR(PDB code 5CRL)[31]. D: Ribbon plot of the TetR dimer N82A in complex with doxycycline-Mg2+(PDB code 6RGX)[25]. E: Ribbon diagram showing the overall structure of the LasR-N-3-oxo-dodecanoyl homoserine lactone homodimer(PDB code 3Ⅸ3)with one monomer colored in light blue and the second colored in purple. The homoserine lactone is shown in a ball-and-stick representation and secondary structure elements are noted on the light-blue monomer[26]
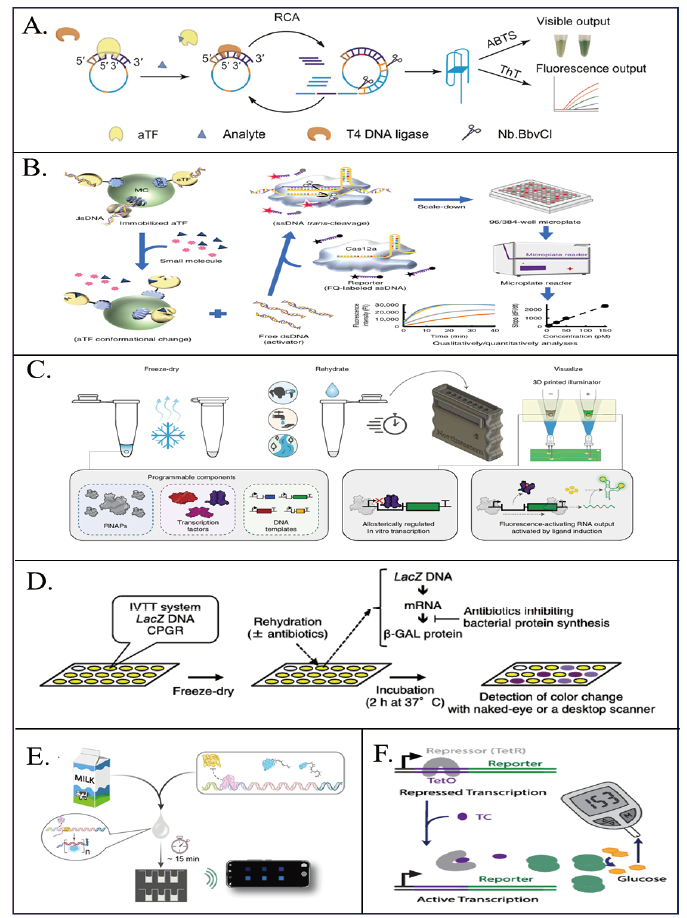
Fig. 3 Application of aTF-based in vivo biosensors in food safety A: Schematic of a biosensor based on aTF coupled with RCA for the detection of small molecules[33]. B: Schematic diagram of a biosensor based on aTF coupled to CRISPR-Cas12a for the detection of small molecules[36]. C: Schematic representation of the RNA Output Sensors Activated by Ligand Induction(ROSALIND)is used to detect contaminants in water in a cell-free in vivo transcription system[39]. D: Schematic of the colorimetric paper-based biosensor for the detection of antibiotics inhibiting bacterial protein synthesis[43]. E: Schematic representation of a smartphone-based aTF bioluminescent sensor for quantification of tetracycline and erythromycin in milk[11]. F: Schematic of a TetO/TetR-based biosensor to detect tetracycline using a glucometer[47]
| 物质类别 Material type | 靶标 Target | 识别元件 Recognition element | 信号输出 Signal output | 检测限 Detection limit | 检测时间 Detection time | 实际样品 Actual sample | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| 重金属离子 Heavy metal ion | Zn2+ | SmtB | 荧光Fluorescence | 2.5 µmol/L | — | 水样Water | [ |
| Cu2+ | CsoR | 荧光Fluorescence | 5 µmol/L | — | 水样Water | [ | |
| Pb2+ | CadC | 荧光Fluorescence | 259 μg/L | — | 水样Water | [ | |
| Cd2+ | CadC | 荧光Fluorescence | 141 μg/L | — | 水样Water | [ | |
| As3+ | ArsR | 比色Colorimetry | 0.5 mmol/L | 3 h | 水样Water | [ | |
| Hg2+ | MerR | 荧光、比色、生物发光Fluorescence,colorimetry, bioluminescence | 1 nmol/L | 30 min | —— | [ | |
| Hg2+ | MerR | 荧光Fluorescence | 0.5 nmol/L | 1 h | 河水、海水和废水River water, seawater, and wastewater | [ | |
| Pb2+ | PbrR | 荧光Fluorescence | 0.1 nmol/L | 1 h | 河水、海水和废水River water, seawater, and wastewater | [ | |
| Hg2+ | MerR | 荧光Fluorescence | 6 μg/L | 1 h | 水样Water | [ | |
| Hg2+ | MerR | 荧光Fluorescence | 1 μg/L | 1 h | 水样Water | [ | |
| As3+ | ArsR | 荧光Fluorescence | 3.65 μg/L | 2.5 h | 水样Water | [ | |
| 农兽药残留 Pesticide and veterinary drug residues | 土霉素Oxytetracycline | OtrR | 荧光Fluorescence | 0.03 nmol/L | — | — | [ |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 18.1 nmol/L | — | — | [ | |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 80 nmol/L | 32 min | — | [ | |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 12.5 nmol/L | 5 min | — | [ | |
| 无水四环素Anhydrotetracycline | TetR | 荧光Fluorescence | 0.5 μmol/L | 60 min | 牛奶Milk | [ | |
| 红霉素Erythromycin | MphR | 荧光Fluorescence | 0.1 μmol/L | 60 min | 牛奶Milk | [ | |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 25 nmol/L | 5-30 min | 水样Water | [ | |
| 红霉素Erythromycin | MphR | 荧光Fluorescence | 100 nmol/L | 5-30 min | 水样Water | [ | |
| 西维因Carbaryl | TtgR | qPCR | — | — | — | [ | |
| 阿特拉津Atrazine | AtzA | 荧光Fluorescence | 20 μmol/L | 1 h | 水样Water | [ | |
| 食品添加剂 Food additives | 4-HBA | HosA | RT-qPCR | 1.12 nmol/L | — | 河水River water | [ |
| 4-HBA | HosA | RPA | 0.005 nmol/L | — | 河水River water | [ | |
| 4-HBA | HosA | RCA | 1.73 nmol/L | — | 河水River water | [ | |
| 乳酸Lactate | STLldR | 荧光Fluorescence | 0.68 μmol/L | — | 细菌发酵样品、酵素和酸奶Bacterial fermentated samples,fermentand yogurt | [ | |
| 乳酸Lactate | LldR | 荧光Fluorescence | 2.34nmol/L | — | — | [ | |
| PHBA | HosA | 荧光Fluorescence | 1.8 nmol/L | 25 min | — | [ | |
| PHBA | HosA | 荧光Fluorescence | 1.35 nmol/L | — | — | [ | |
| PHBA | HosA | 比色Colorimetry | 2.55 nmol/L | — | — | [ | |
| 苯甲酸Benzoic acid | BenR | 荧光Fluorescence | — | ~1 h | 饮料Beverages | [ | |
| 苯甲酸Benzoic acid | BenR | 荧光Fluorescence | 21 702.843 nmol/L | — | 水样Water | [ | |
| 食源性病原体 Foodborne pathogen | 铜绿假单胞菌Pseudomonas aeruginosa | LasRV | 荧光Fluorescence | 4.9 nmol/L | — | 痰样Sputum | [ |
| 铜绿假单胞菌Pseudomonas aeruginosa | LasR | 荧光Fluorescence | 10 nmol/L | — | — | [ |
Table 1 Applications of aTF biosensors in food safety
| 物质类别 Material type | 靶标 Target | 识别元件 Recognition element | 信号输出 Signal output | 检测限 Detection limit | 检测时间 Detection time | 实际样品 Actual sample | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| 重金属离子 Heavy metal ion | Zn2+ | SmtB | 荧光Fluorescence | 2.5 µmol/L | — | 水样Water | [ |
| Cu2+ | CsoR | 荧光Fluorescence | 5 µmol/L | — | 水样Water | [ | |
| Pb2+ | CadC | 荧光Fluorescence | 259 μg/L | — | 水样Water | [ | |
| Cd2+ | CadC | 荧光Fluorescence | 141 μg/L | — | 水样Water | [ | |
| As3+ | ArsR | 比色Colorimetry | 0.5 mmol/L | 3 h | 水样Water | [ | |
| Hg2+ | MerR | 荧光、比色、生物发光Fluorescence,colorimetry, bioluminescence | 1 nmol/L | 30 min | —— | [ | |
| Hg2+ | MerR | 荧光Fluorescence | 0.5 nmol/L | 1 h | 河水、海水和废水River water, seawater, and wastewater | [ | |
| Pb2+ | PbrR | 荧光Fluorescence | 0.1 nmol/L | 1 h | 河水、海水和废水River water, seawater, and wastewater | [ | |
| Hg2+ | MerR | 荧光Fluorescence | 6 μg/L | 1 h | 水样Water | [ | |
| Hg2+ | MerR | 荧光Fluorescence | 1 μg/L | 1 h | 水样Water | [ | |
| As3+ | ArsR | 荧光Fluorescence | 3.65 μg/L | 2.5 h | 水样Water | [ | |
| 农兽药残留 Pesticide and veterinary drug residues | 土霉素Oxytetracycline | OtrR | 荧光Fluorescence | 0.03 nmol/L | — | — | [ |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 18.1 nmol/L | — | — | [ | |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 80 nmol/L | 32 min | — | [ | |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 12.5 nmol/L | 5 min | — | [ | |
| 无水四环素Anhydrotetracycline | TetR | 荧光Fluorescence | 0.5 μmol/L | 60 min | 牛奶Milk | [ | |
| 红霉素Erythromycin | MphR | 荧光Fluorescence | 0.1 μmol/L | 60 min | 牛奶Milk | [ | |
| 四环素Tetracycline | TetR | 荧光Fluorescence | 25 nmol/L | 5-30 min | 水样Water | [ | |
| 红霉素Erythromycin | MphR | 荧光Fluorescence | 100 nmol/L | 5-30 min | 水样Water | [ | |
| 西维因Carbaryl | TtgR | qPCR | — | — | — | [ | |
| 阿特拉津Atrazine | AtzA | 荧光Fluorescence | 20 μmol/L | 1 h | 水样Water | [ | |
| 食品添加剂 Food additives | 4-HBA | HosA | RT-qPCR | 1.12 nmol/L | — | 河水River water | [ |
| 4-HBA | HosA | RPA | 0.005 nmol/L | — | 河水River water | [ | |
| 4-HBA | HosA | RCA | 1.73 nmol/L | — | 河水River water | [ | |
| 乳酸Lactate | STLldR | 荧光Fluorescence | 0.68 μmol/L | — | 细菌发酵样品、酵素和酸奶Bacterial fermentated samples,fermentand yogurt | [ | |
| 乳酸Lactate | LldR | 荧光Fluorescence | 2.34nmol/L | — | — | [ | |
| PHBA | HosA | 荧光Fluorescence | 1.8 nmol/L | 25 min | — | [ | |
| PHBA | HosA | 荧光Fluorescence | 1.35 nmol/L | — | — | [ | |
| PHBA | HosA | 比色Colorimetry | 2.55 nmol/L | — | — | [ | |
| 苯甲酸Benzoic acid | BenR | 荧光Fluorescence | — | ~1 h | 饮料Beverages | [ | |
| 苯甲酸Benzoic acid | BenR | 荧光Fluorescence | 21 702.843 nmol/L | — | 水样Water | [ | |
| 食源性病原体 Foodborne pathogen | 铜绿假单胞菌Pseudomonas aeruginosa | LasRV | 荧光Fluorescence | 4.9 nmol/L | — | 痰样Sputum | [ |
| 铜绿假单胞菌Pseudomonas aeruginosa | LasR | 荧光Fluorescence | 10 nmol/L | — | — | [ |
| [1] | Wang PL, Xie LH, Joseph EA, et al. Metal-organic frameworks for food safety[J]. Chem Rev, 2019, 119(18): 10638-10690. |
| [2] | 张秋月. 土壤污染治理问题研究——以镉大米事件为视角[J]. 法制与社会, 2017(3): 77-78. |
| Zhang QY. Research on soil pollution control issues - taking the cadmium rice incident as a perspective[J]. Leg Syst Soc, 2017(3): 77-78. | |
| [3] |
Yamaguchi T, Okihashi M, Harada K, et al. Detection of antibiotics in chicken eggs obtained from supermarkets in Ho Chi Minh City, Vietnam[J]. J Environ Sci Health B, 2017, 52(6): 430-433.
doi: 10.1080/03601234.2017.1293457 pmid: 28281880 |
| [4] | 李航, 刘议蔧, 余晓琴, 等. “穿透式” 监管下的酱油、食醋食品安全问题及对策研究[J]. 食品工业, 2024, 45(4): 323-326. |
| Li H, Liu YH, Yu XQ, et al. Study on food safety problems and countermeasures of soy sauce and vinegar from the “penetrating” supervision[J]. Food Ind, 2024, 45(4): 323-326. | |
| [5] | Griesche C, Baeumner AJ. Biosensors to support sustainable agriculture and food safety[J]. Trac Trends Anal Chem, 2020, 128: 115906. |
| [6] |
Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes[J]. Trends Microbiol, 2005, 13(2): 52-56.
pmid: 15680762 |
| [7] |
Libis V, Delépine B, Faulon JL. Sensing new chemicals with bacterial transcription factors[J]. Curr Opin Microbiol, 2016, 33: 105-112.
doi: S1369-5274(16)30094-7 pmid: 27472026 |
| [8] |
Baksh KA, Zamble DB. Allosteric control of metal-responsive transcriptional regulators in bacteria[J]. J Biol Chem, 2020, 295(6): 1673-1684.
doi: 10.1074/jbc.REV119.011444 pmid: 31857375 |
| [9] | Zhang YK, Zhao C, Bi HX, et al. A cell-free paper-based biosensor dependent on allosteric transcription factors(aTFs)for on-site detection of harmful metals Hg2+ and Pb2+ in water[J]. J Hazard Mater, 2022, 438: 129499. |
| [10] |
Zhang P, Feng HB, Yang JZ, et al. Detection of inorganic ions and organic molecules with cell-free biosensing systems[J]. J Biotechnol, 2019, 300: 78-86.
doi: S0168-1656(19)30175-0 pmid: 31141711 |
| [11] | Zhang R, Wang Y, Deng HF, et al. Fast and bioluminescent detection of antibiotic contaminants by on-demand transcription of RNA scaffold arrays[J]. Anal Chim Acta, 2023, 1273: 341538. |
| [12] | Li SS, Zhou L, Yao YP, et al. A platform for the development of novel biosensors by configuring allosteric transcription factor recognition with amplified luminescent proximity homogeneous assays[J]. Chem Commun, 2016, 53(1): 99-102. |
| [13] |
Silverman AD, Akova U, Alam KK, et al. Design and optimization of a cell-free atrazine biosensor[J]. ACS Synth Biol, 2020, 9(3): 671-677.
doi: 10.1021/acssynbio.9b00388 pmid: 32078765 |
| [14] | Yao YP, Li SS, Cao JQ, et al. Development of small molecule biosensors by coupling the recognition of the bacterial allosteric transcription factor with isothermal strand displacement amplification[J]. Chem Commun, 2018, 54(38): 4774-4777. |
| [15] | Xiao D, Hu CX, Xu XZ, et al. A d, l-lactate biosensor based on allosteric transcription factor LldR and amplified luminescent proximity homogeneous assay[J]. Biosens Bioelectron, 2022, 211: 114378. |
| [16] | Voyvodic PL, Pandi A, Koch M, et al. Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors[J]. Nat Commun, 2019, 10(1): 1697. |
| [17] | Jong S. On a robust, sensitive cell-free method for Pseudomonas sensing and quantification in microfluidic templated hydrogels[J]. Micromachines, 2019, 10(8): 506. |
| [18] | Liu YE, Guo MZ, Du RX, et al. A gas reporting whole-cell microbial biosensor system for rapid on-site detection of mercury contamination in soils[J]. Biosens Bioelectron, 2020, 170: 112660. |
| [19] |
Li SS, Li ZL, Tan GY, et al. in vivo allosteric transcription factor-based biosensing[J]. Trends Biotechnol, 2023, 41(8): 1080-1095.
doi: 10.1016/j.tibtech.2023.03.001 pmid: 36967257 |
| [20] | Wang GH, Wang F, Huang Q, et al. Understanding transcription factor regulation by integrating gene expression and DNase I hypersensitive sites[J]. Biomed Res Int, 2015, 2015: 757530. |
| [21] | Ding NN, Zhou SH, Deng Y. Transcription-factor-based biosensor engineering for applications in synthetic biology[J]. ACS Synth Biol, 2021, 10(5): 911-922. |
| [22] |
Tulin G, Figueroa NR, Checa SK, et al. The multifarious MerR family of transcriptional regulators[J]. Mol Microbiol, 2024, 121(2): 230-242.
doi: 10.1111/mmi.15212 pmid: 38105009 |
| [23] | Bhukya H, Anand R. TetR regulators: a structural and functional perspective[J]. J Indian Inst Sci, 2017, 97(2): 245-259. |
| [24] | Deochand DK, Grove A. MarR family transcription factors: dynamic variations on a common scaffold[J]. Crit Rev Biochem Mol Biol, 2017, 52(6): 595-613. |
| [25] | Palm GJ, Buchholz I, Werten S, et al. Thermodynamics, cooperativity and stability of the tetracycline repressor(TetR)upon tetracycline binding[J]. Biochim Biophys Acta Proteins Proteom, 2020, 1868(6): 140404. |
| [26] | Zou YZ, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor[J]. Chem Biol, 2009, 16(9): 961-970. |
| [27] |
Kasey CM, Zerrad M, Li YW, et al. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology[J]. ACS Synth Biol, 2018, 7(1): 227-239.
doi: 10.1021/acssynbio.7b00287 pmid: 28950701 |
| [28] |
Machado LFM, Currin A, Dixon N. Directed evolution of the PcaV allosteric transcription factor to generate a biosensor for aromatic aldehydes[J]. J Biol Eng, 2019, 13: 91.
doi: 10.1186/s13036-019-0214-z pmid: 31798685 |
| [29] |
Bai DY, Ding DQ, Li JL, et al. Pinpointing the L-phenylalanine binding sites of TyrR using biosensors and computer-aided simulation[J]. Biotechnol Lett, 2019, 41(3): 401-408.
doi: 10.1007/s10529-019-02645-x pmid: 30680497 |
| [30] | Liang YY, Luo J, Yang CH, et al. Directed evolution of the PobR allosteric transcription factor to generate a biosensor for 4-hydroxymandelic acid[J]. World J Microbiol Biotechnol, 2022, 38(6): 104. |
| [31] |
Wang D, Huang SQ, Liu PY, et al. Structural analysis of the Hg(II)-regulatory protein Tn501 MerR from Pseudomonas aeruginosa[J]. Sci Rep, 2016, 6: 33391.
doi: 10.1038/srep33391 pmid: 27641146 |
| [32] | Liu RN, Liu X, Yang H, et al. A cell-free biosensor based on strand displacement amplification and hybridization chain reaction for fluorescence detection of tetracycline[J]. Microchem J, 2023, 185: 108239. |
| [33] | Cao JQ, Yao YP, Fan KQ, et al. Harnessing a previously unidentified capability of bacterial allosteric transcription factors for sensing diverse small molecules in vivo[J]. Sci Adv, 2018, 4(11): eaau4602. |
| [34] | Zhong JL, Zhao XH. Isothermal amplification technologies for the detection of foodborne pathogens[J]. Food Anal Meth, 2018, 11(6): 1543-1560. |
| [35] | Iwasaki RS, Batey RT. SPRINT: a Cas13a-based platform for detection of small molecules[J]. Nucleic Acids Res, 2020, 48(17): e101. |
| [36] | Liang MD, Li ZL, Wang WS, et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules[J]. Nat Commun, 2019, 10(1): 3672. |
| [37] |
Özyurt C, Üstükarcı H, Evran S, et al. MerR-fluorescent protein chimera biosensor for fast and sensitive detection of Hg2+ in drinking water[J]. Biotechnol Appl Biochem, 2019, 66(5): 731-737.
doi: 10.1002/bab.1805 pmid: 31411759 |
| [38] | Gräwe A, Dreyer A, Vornholt T, et al. A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs[J]. PLoS One, 2019, 14(3): e0210940. |
| [39] | Jung JK, Alam KK, Verosloff MS, et al. Cell-free biosensors for rapid detection of water contaminants[J]. Nat Biotechnol, 2020, 38(12): 1451-1459. |
| [40] | Nguyen TT, Chern M, Baer RC, et al. A Förster resonance energy transfer-based ratiometric sensor with the allosteric transcription factor TetR[J]. Small, 2020, 16(17): e1907522. |
| [41] | Chen MF, Nguyen TT, Varongchayakul N, et al. Surface immobilized nucleic acid-transcription factor quantum dots for biosensing[J]. Adv Healthc Mater, 2020, 9(17): e2000403. |
| [42] |
Lin XM, Li YT, Li ZX, et al. Portable environment-signal detection biosensors with cell-free synthetic biosystems[J]. RSC Adv, 2020, 10(64): 39261-39265.
doi: 10.1039/d0ra05293k pmid: 35518409 |
| [43] |
Duyen TTM, Matsuura H, Ujiie K, et al. Paper-based colorimetric biosensor for antibiotics inhibiting bacterial protein synthesis[J]. J Biosci Bioeng, 2017, 123(1): 96-100.
doi: S1389-1723(16)30186-4 pmid: 27514909 |
| [44] | Brodl E, Winkler A, Macheroux P. Molecular mechanisms of bacterial bioluminescence[J]. Comput Struct Biotechnol J, 2018, 16: 551-564. |
| [45] |
Pellinen T, Huovinen T, Karp M. A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter[J]. Anal Biochem, 2004, 330(1): 52-57.
pmid: 15183761 |
| [46] |
Lopreside A, Wan XY, Michelini E, et al. Comprehensive profiling of diverse genetic reporters with application to whole-cell and cell-free biosensors[J]. Anal Chem, 2019, 91(23): 15284-15292.
doi: 10.1021/acs.analchem.9b04444 pmid: 31690077 |
| [47] | Amalfitano E, Karlikow M, Norouzi M, et al. A glucose meter interface for point-of-care gene circuit-based diagnostics[J]. Nat Commun, 2021, 12(1): 724. |
| [48] | Sankar K, Baer R, Grazon C, et al. An allosteric transcription factor DNA-binding electrochemical biosensor for progesterone[J]. ACS Sens, 2022, 7(4): 1132-1137. |
| [49] |
Yao YP, Li SS, Cao JQ, et al. A novel signal transduction system for development of uric acid biosensors[J]. Appl Microbiol Biotechnol, 2018, 102(17): 7489-7497.
doi: 10.1007/s00253-018-9056-8 pmid: 29961098 |
| [50] | Wang T, Lu Y. Advances, challenges and future trends of cell-free transcription-translation biosensors[J]. Biosensors, 2022, 12(5): 318. |
| [51] | Bi HX, Zhao C, Zhang YK, et al. IVT cell-free biosensors for tetracycline and macrolide detection based on allosteric transcription factors(aTFs)[J]. Anal Methods, 2022, 14(44): 4545-4554. |
| [52] | Kudo A, Fujikawa Y, Miyahara S, et al. Lessons from Minamata mercury pollution, Japan - after a continuous 22 years of observation[J]. Water Sci Technol, 1998, 38(7): 187-193. |
| [53] |
Gupta S, Sarkar S, Katranidis A, et al. Development of a cell-free optical biosensor for detection of a broad range of mercury contaminants in water: a plasmid DNA-based approach[J]. ACS Omega, 2019, 4(5): 9480-9487.
doi: 10.1021/acsomega.9b00205 pmid: 31460039 |
| [54] | Wang XY, Zhu KL, Chen DD, et al. Monitoring arsenic using genetically encoded biosensors in vivo: the role of evolved regulatory genes[J]. Ecotoxicol Environ Saf, 2021, 207: 111273. |
| [55] | Wang JS, Davidson JL, Kaur S, et al. Paper-based biosensors for the detection of nucleic acids from pathogens[J]. Biosensors, 2022, 12(12): 1094. |
| [56] | Jia M, Zhongbo E, Zhai F, et al. Rapid multi-residue detection methods for pesticides and veterinary drugs[J]. Molecules, 2020, 25(16): 3590. |
| [57] | Chern M, Garden PM, Baer RC, et al. Transcription factor based small-molecule sensing with a rapid cell phone enabled fluorescent bead assay[J]. Angew Chem Int Ed Engl, 2020, 59(48): 21597-21602. |
| [58] |
Rodríguez-Serrano AF, Hsing IM. Allosteric regulation of DNA circuits enables minimal and rapid biosensors of small molecules[J]. ACS Synth Biol, 2021, 10(2): 371-378.
doi: 10.1021/acssynbio.0c00545 pmid: 33481567 |
| [59] |
Mahas A, Wang QC, Marsic T, et al. Development of Cas12a-based cell-free small-molecule biosensors via allosteric regulation of CRISPR array expression[J]. Anal Chem, 2022, 94(11): 4617-4626.
doi: 10.1021/acs.analchem.1c04332 pmid: 35266687 |
| [60] |
Chen W, Zhang XX, Xiong DD, et al. Engineering the effector specificity of regulatory proteins for the in vivo detection of biomarkers and pesticide residues[J]. Appl Microbiol Biotechnol, 2019, 103(7): 3205-3213.
doi: 10.1007/s00253-019-09679-1 pmid: 30770965 |
| [61] | Warner JO. Artificial food additives: hazardous to long-term health[J]. Arch Dis Child, 2024: archdischild-archdisc2023-326565. |
| [62] | Marques C, Wojeicchowski JP, Cardoso T, et al. Lactobionic acid as a suitable food preservative for yacon juice[J]. Innov Food Sci Emerg Technol, 2020, 64: 102400. |
| [63] | Xu XZ, Xu R, Hou S, et al. A selective fluorescent l-lactate biosensor based on an l-lactate-specific transcription regulator and Förster resonance energy transfer[J]. Biosensors, 2022, 12(12): 1111. |
| [64] | Saravanan A, Kumar PS, Hemavathy RV, et al. Methods of detection of food-borne pathogens: a review[J]. Environ Chem Lett, 2021, 19(1): 189-207. |
| [65] | Verbeke F, De Craemer S, Debunne N, et al. Peptides as quorum sensing molecules: measurement techniques and obtained levels in vivo and In vivo[J]. Front Neurosci, 2017, 11: 183. |
| [66] | Wen KY, Cameron L, Chappell J, et al. A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa-infected respiratory samples[J]. ACS Synth Biol, 2017, 6(12): 2293-2301. |
| [1] | ZHANG Di, JU Rui, LI Li-mei, WANG Yu-qian, CHEN Rui, WANG Xin-yi. Application of Transcription Factor-based Biosensors in Environmental Analysis [J]. Biotechnology Bulletin, 2024, 40(6): 114-125. |
| [2] | WANG Meng-ya, LIU Jia-qi, JIANG Hai-lin, LI Jing-hua, ZHAO Chun-yan, HUANG Hong-lan. Biological Characteristics and Application of Enteroinvasive Escherichia coli Phage DK-13 [J]. Biotechnology Bulletin, 2024, 40(3): 296-304. |
| [3] | YU Yong-xia, ZHU Ning, LIU Guang-min, ZHU Long-jiao, XU Wen-tao. Research Progress in Nucleic Acid Molecular Diagnostic Technology for Mycoplasma pneumoniae [J]. Biotechnology Bulletin, 2024, 40(12): 72-83. |
| [4] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [5] | LI Ren-han, ZHANG Le-le, LIU Chun-li, LIU Xiu-xia, BAI Zhong-hu, YANG Yan-kun, LI Ye. Development of an L-tryptophan Biosensor Based on the Violacein Biosynthesis Pathway [J]. Biotechnology Bulletin, 2023, 39(10): 80-92. |
| [6] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| [7] | ZHANG Ya-han, ZHU Li-xia, HU Jing, ZHU Ya-jing, ZHANG Xue-jing, CAO Ye-zhong. Opportunities and Challenges of Glyphosate in the Application of Biotechnology Breeding in China [J]. Biotechnology Bulletin, 2022, 38(11): 1-9. |
| [8] | WANG Peng-fei, YANG Min, ZHU Long-jiao, XU Wen-tao. Advances in Biosensors Based on Platinum Nanoclusters [J]. Biotechnology Bulletin, 2021, 37(12): 235-242. |
| [9] | ZHAO Ying, WANG Nan, LU An-xiang, FENG Xiao-yuan, GUO Xiao-jun, LUAN Yun-xia. Application in the Detection of Fungal Toxins by Nucleic Acid Aptamer Lateral Flow Chromatography Analysis Technique [J]. Biotechnology Bulletin, 2020, 36(8): 217-227. |
| [10] | FANG Shun-yan, SONG Dan, LIU Yan-ping, XU Wen-juan, LIU Jia-yao, HAN Xiang-zhi, LONG Feng. Study on Evanescent Wave Fluorescence Aptasensor for Direct and Rapid Detection of Escherichia coli O157∶H7 [J]. Biotechnology Bulletin, 2020, 36(7): 228-234. |
| [11] | YE Jian-wen, CHEN Jiang-nan, ZHANG Xu, Wu Fu-qing, CHEN Guo-qiang. Dynamic Control:An Efficient Strategy for Metabolically Engineering Microbial Cell Factories [J]. Biotechnology Bulletin, 2020, 36(6): 1-12. |
| [12] | YANG Min, LI Shu-ting, YANG Wen-ping, LI Xiang-yang, XU Wen-tao. Research Progress on Functional Nucleic Acid Biosensors Mediated by DNA/Silver Nanoclusters [J]. Biotechnology Bulletin, 2020, 36(6): 245-254. |
| [13] | LIU Su-yue, TIAN Jing-jing, TIAN Hong-tao, XU Wen-tao. Terbium(III)and Its Complexes:from Luminescent Properties to Sensing and Bioimaging Applications [J]. Biotechnology Bulletin, 2020, 36(4): 192-207. |
| [14] | SUN Yu-ge, LI Chen-wei, DU Zai-hui, XU Wen-tao. Research Progress on FEN1-mediated Functional Nucleic Acid Biosensors [J]. Biotechnology Bulletin, 2020, 36(4): 208-224. |
| [15] | WANG Qi, YAN Chun-lei, GAO Hong-wei, WU Wei, YANG Qing-li. Research Progress of DNA Aptasensors for Foodborne Pathogen Detection [J]. Biotechnology Bulletin, 2020, 36(11): 245-258. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||