








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 219-237.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1195
Previous Articles Next Articles
HU Yong-bo1,2( ), LEI Yu-tian1,2, YANG Yong-sen1,2, CHEN Xin1,2, LIN Huang-fang1,2, LIN Bi-ying1,2, LIU Shuang1,2, BI Ge1,2, SHEN Bao-ying1,2(
), LEI Yu-tian1,2, YANG Yong-sen1,2, CHEN Xin1,2, LIN Huang-fang1,2, LIN Bi-ying1,2, LIU Shuang1,2, BI Ge1,2, SHEN Bao-ying1,2( )
)
Received:2023-12-15
Online:2024-06-26
Published:2024-05-14
Contact:
SHEN Bao-ying
E-mail:194997643@qq.com;shenby889@foxmail.com
HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch.[J]. Biotechnology Bulletin, 2024, 40(6): 219-237.
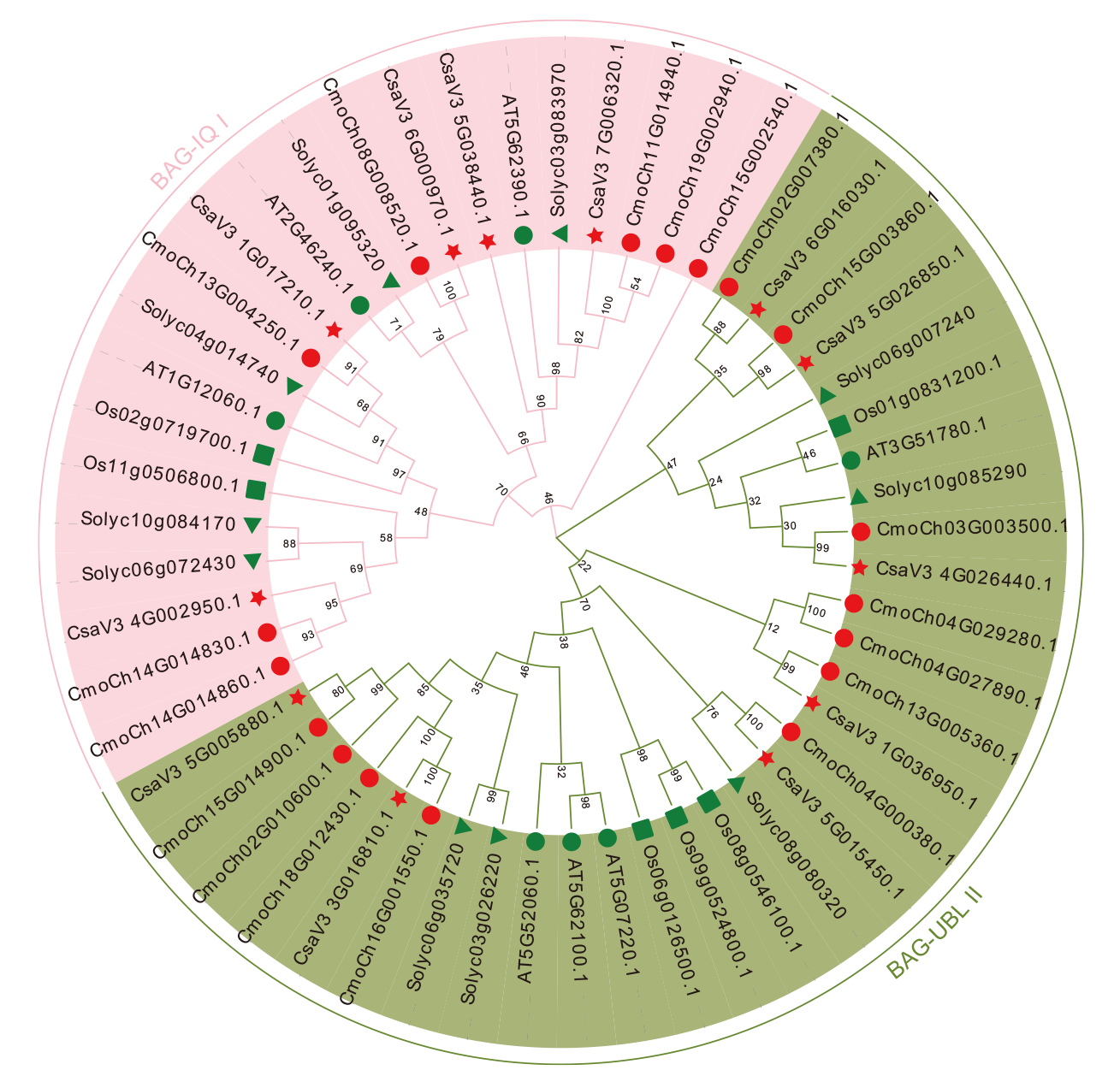
Fig. 2 Phylogenetic analysis of the BAG gene family in C. sativus L., C. moschata Duch., Arabidopsis, O. sativa L. and S. lyco-persicum L. : C. sativus L.. : C. moschata Duch.. : Arabidopsis. : O. sativa L.. : S. lycopersicum L
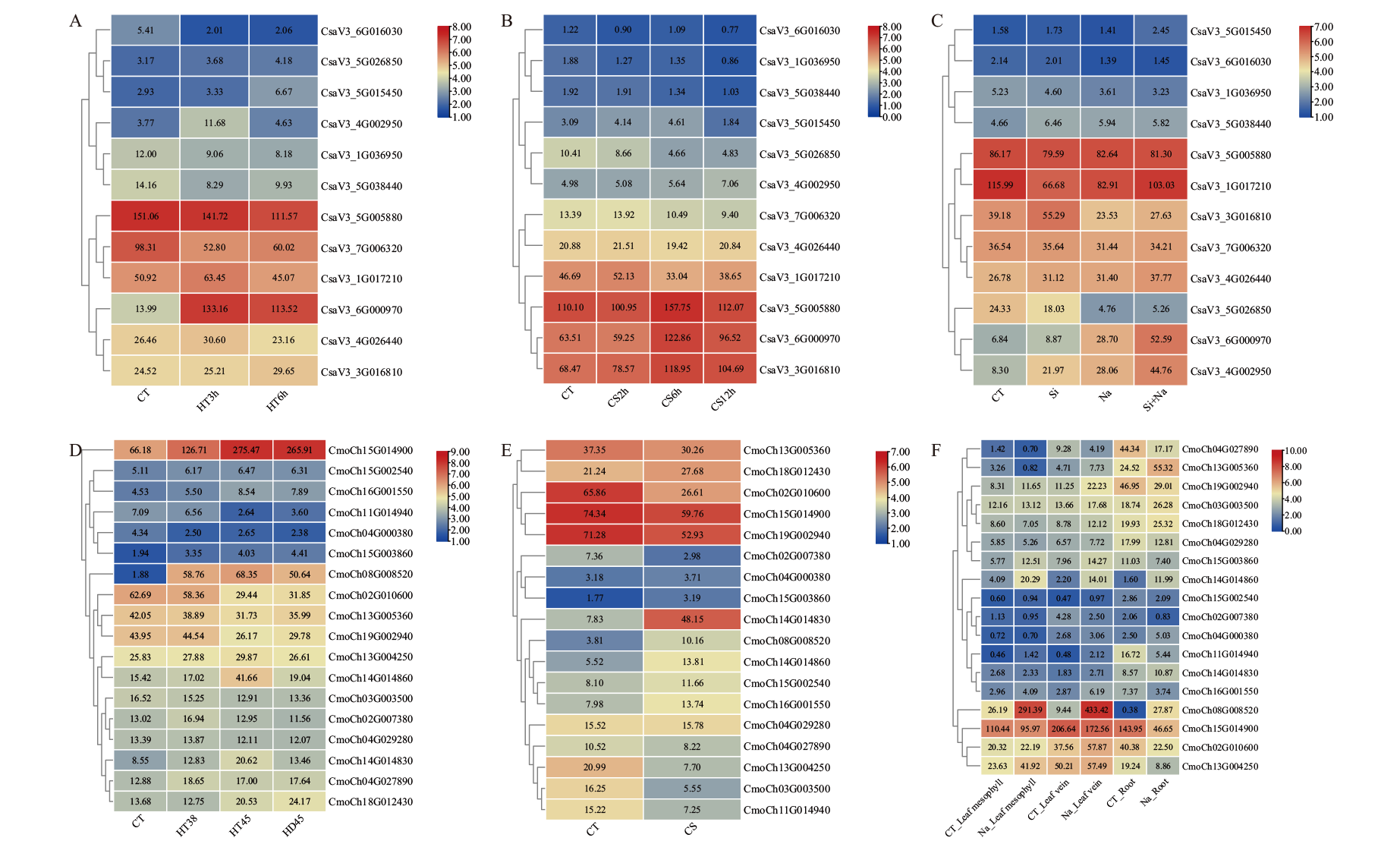
Fig. 7 Expressions of BAG gene family in C. sativus L. and C. moschata Duch. under abiotic stress A: High temperature stress in C. sativus L. (HT3h, HT6h: High temperature treatment for 3 and 6 h). B: Low temperature stress in C. sativus L. (CS2h, CS6h and CS12h: Low temperature treatment for 2, 6 and 12 h). C: Salt and silicon stress in C. sativus L. (Na: Salt stress treatment; Si: silicon stress treatment; Si+Na: silicon and salt stress treatment). D: High temperature stress in C. moschata Duch. (HT38: 38°C high temperature treatment for 3 h; HD45: 38°C high temperature adaptation for 3 h, recovered at 25°C for 5 h, and then excited at 45°C for 3 h; HT45: high temperature treatment at 45°C for 3 h). E: Low temperature stress in C. moschata Duch. (CS: Low temperature treatment). F: Salt stress in C. moschata Duch. (CT_Leaf mesophyll, CT_Leaf vein, and CT_Root: Control treatment of mesophyll, leaf veins, and roots; Na_Leaf mesophyll, Na_Leaf vein and, Na_Root: salt stress treatment of mesophyll, leaf veins, and roots. CT: Control. The data in the boxes indicate original FPKM value
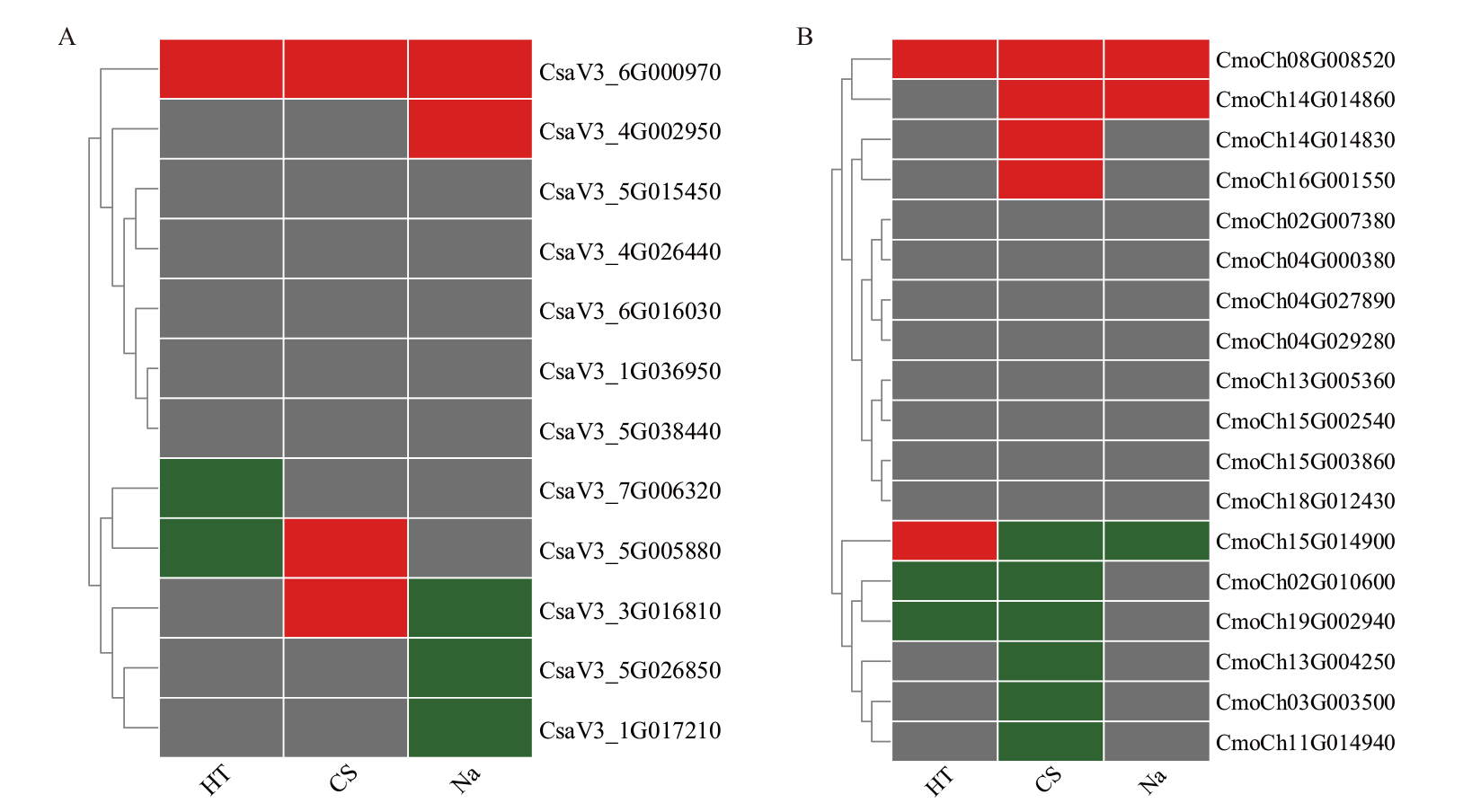
Fig. 8 Expression patterns of BAG family genes in C. sativus L. (A) and C. moschata Duch. (B) under stress HT: High temperature stress. CS: Low temperature stress. Na: Salt stress. The red color indicates the up-regulation of the expression, the green color indicates the down-regulation of the expression, and the gray color indicates the expression is no significant difference in the expression, the same below
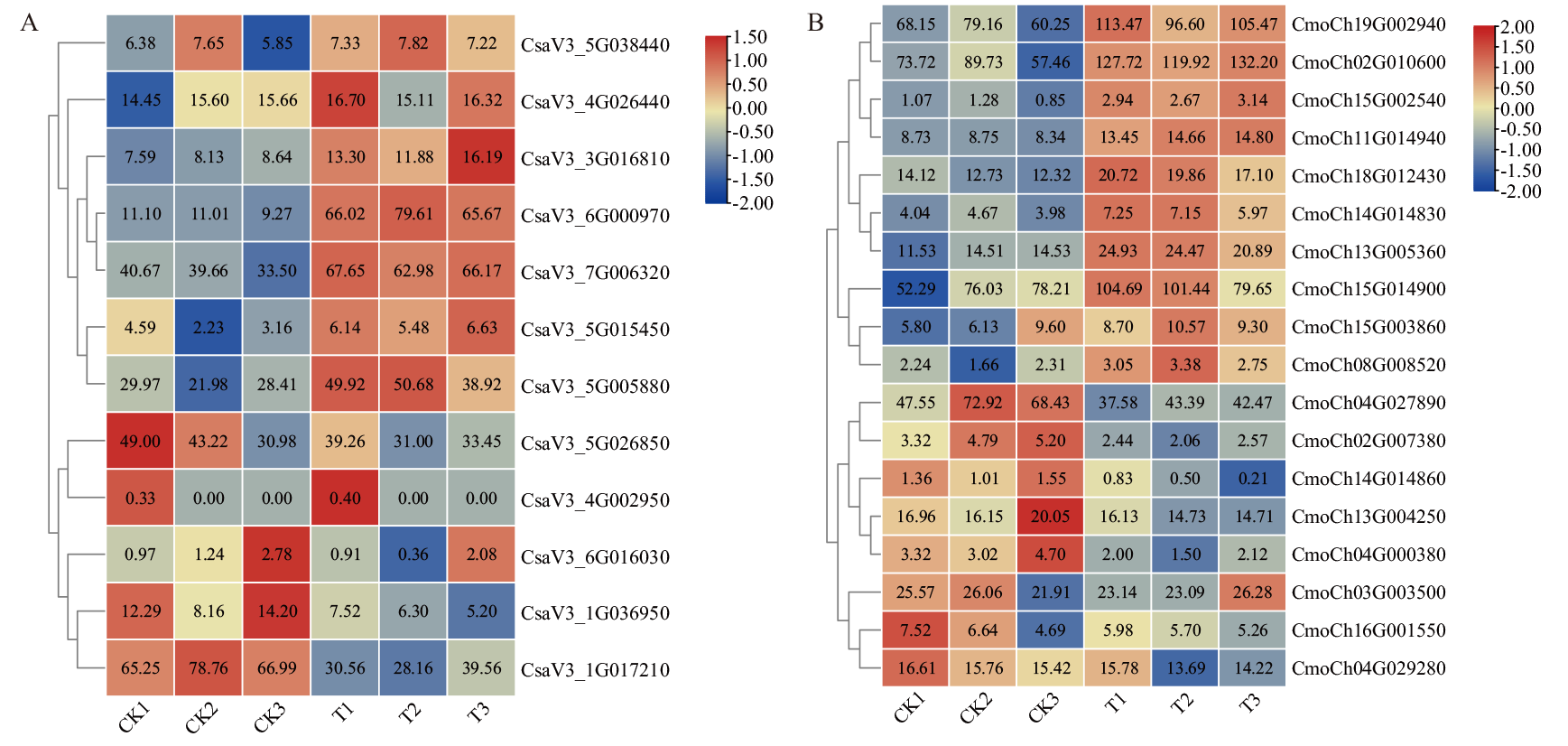
Fig. 9 Expression heat map of BAG gene family in C. sativus L. (A) and C. moschata Duch. (B) in grafting healing CK1, CK2, CK3: Dark control group. T1, T2, T3: Lighting treatment group. The same below
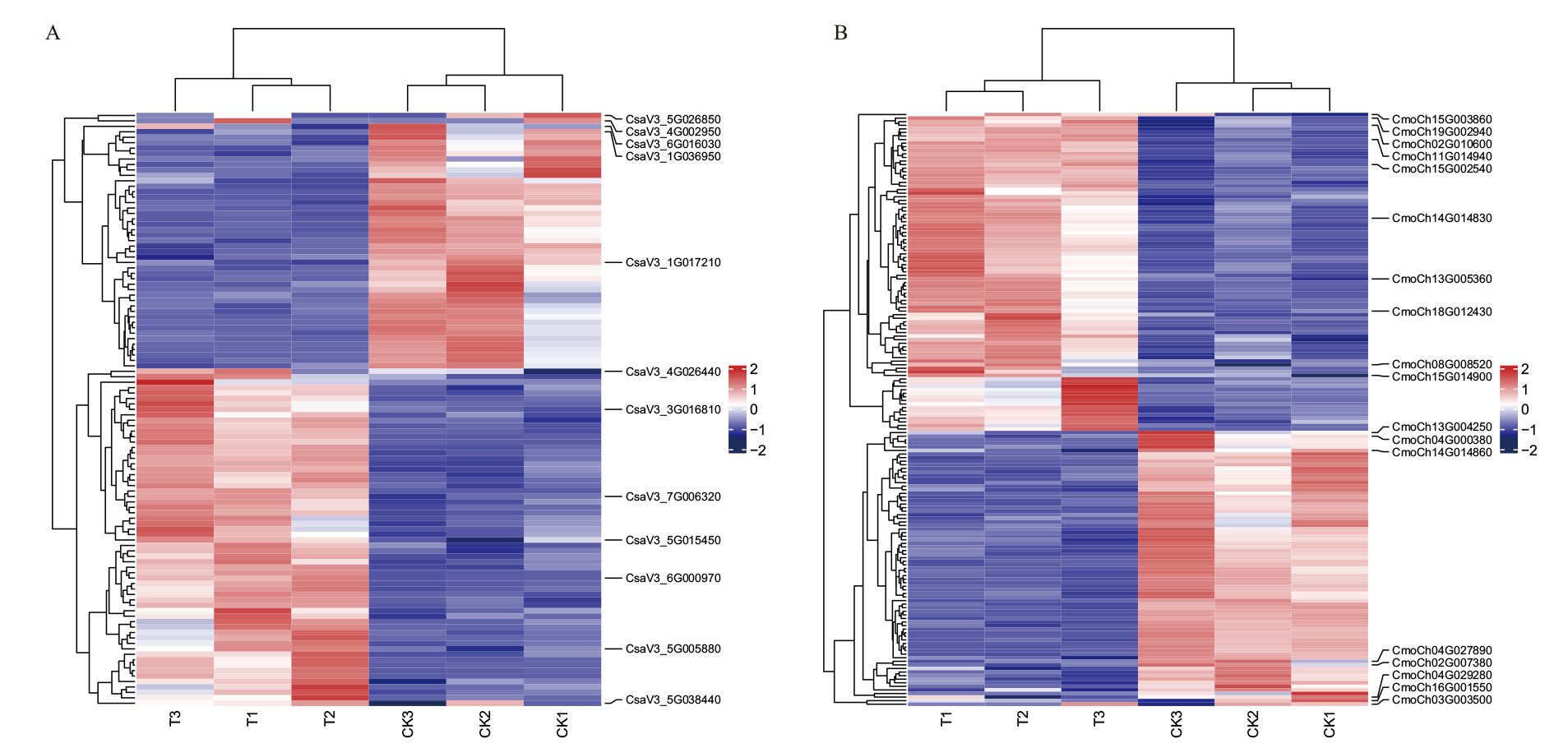
Fig. 10 Co-expression patterns of BAG family genes and differential genes in glucose metabolism pathway in C. sativus L. (A) and C. moschata Duch. (B)
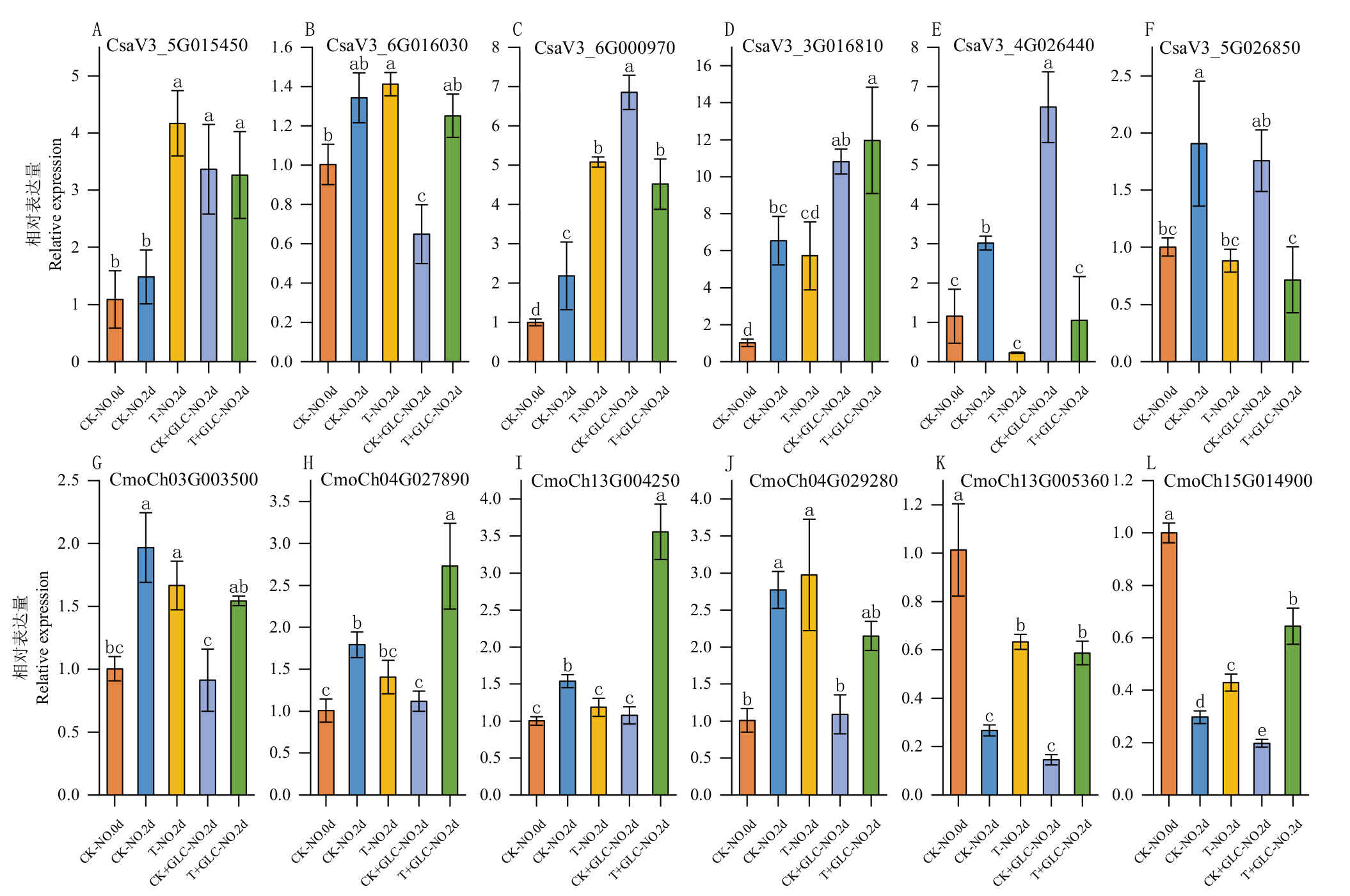
Fig. 11 RT-qPCR validation of BAG gene family in C. sativus L. (A-F) and C. moschata Duch. (G-L) under light and exogenous sugar treatments CK-NO.0d: 0 d dark control group. CK-NO. 2d: 2 d dark control group. T-NO.2d: 2 d lighting treatment group. CK+GLU-NO.2d: 2 d dark + sugar treatment group. T+GLU-NO.2d: 2 d light + sugar treatment group. Different letters indicate significant difference at P<0.05 level
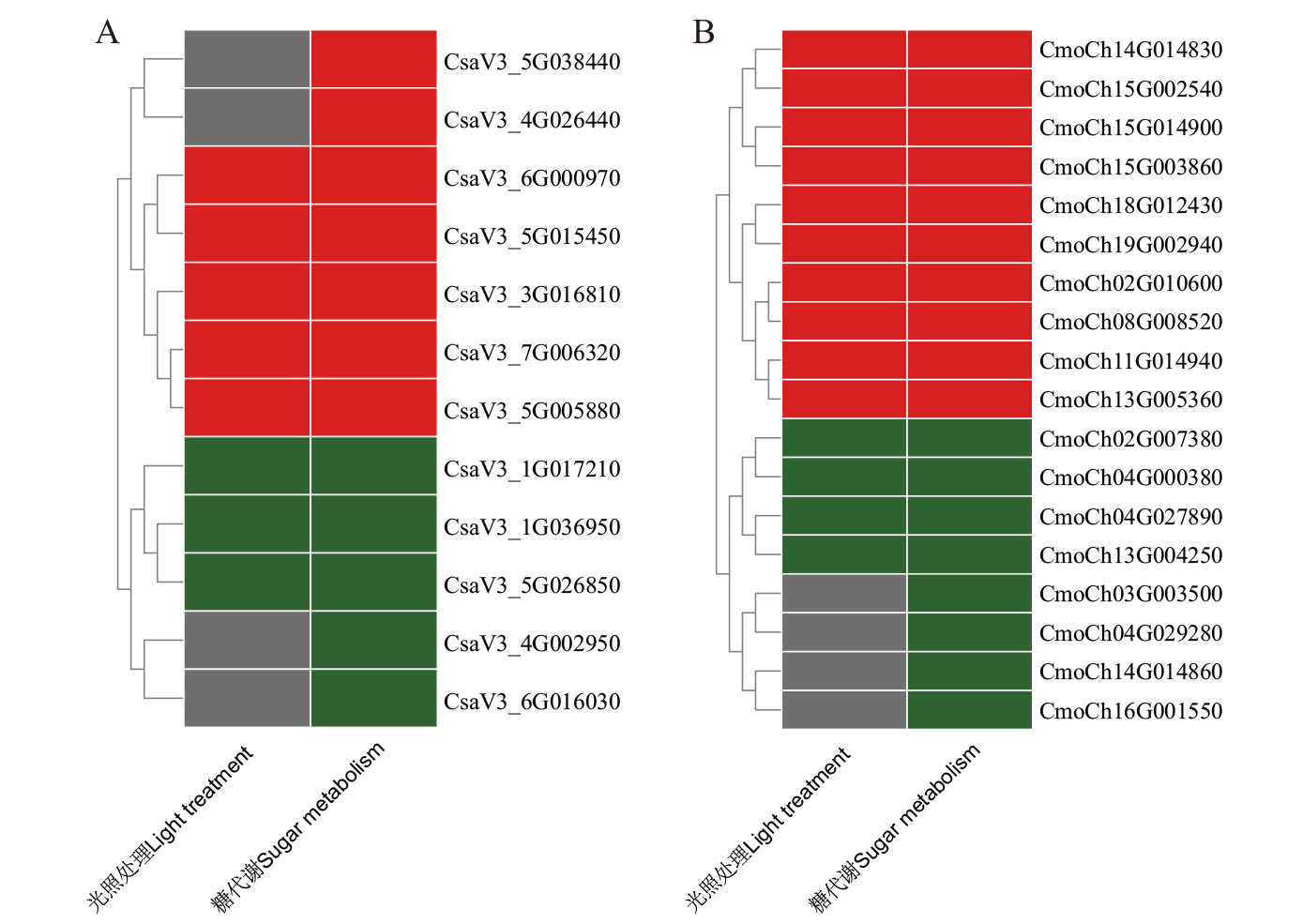
Fig. 12 Expression patterns of BAG family genes in C. sativus L. (A) and C. moschata Duch. (B) during grafting healing Light: Lighting treatment. Sugar: Co-expression of sugar metabolism
| [1] |
Thanthrige N, J ain S, Bhowmik SD, et al. Centrality of BAGs in plant PCD, stress responses, and host defense[J]. Trends Plant Sci, 2020, 25(11): 1131-1140.
doi: 10.1016/j.tplants.2020.04.012 pmid: 32467063 |
| [2] | Huang SW, Li RQ, Z hang ZH, et al. The genome of the cucumber, Cucumis sativus L.[J]. Nat Genet, 2009, 41(12): 1275-1281. |
| [3] |
Briknarová K, Takayama S, Brive L, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein[J]. Nat Struct Biol, 2001, 8(4): 349-352.
pmid: 11276257 |
| [4] |
Takayama S, Sato T, Krajewski S, et al. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity[J]. Cell, 1995, 80(2): 279-284.
pmid: 7834747 |
| [5] |
Doukhanina EV, Chen SR, van der Zalm E, et al. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana[J]. J Biol Chem, 2006, 281(27): 18793-18801.
doi: 10.1074/jbc.M511794200 pmid: 16636050 |
| [6] | Poulsen EG, K ampmeyer C, Kriegenburg F, et al. UBL/BAG-domain co-chaperones cause cellular stress upon overexpression through constitutive activation of Hsf1[J]. Cell Stress & Chaperones, 2017, 22(1): 143-154. |
| [7] |
Yang TB, Poovaiah BW. Calcium/calmodulin-mediated signal network in plants[J]. Trends Plant Sci, 2003, 8(10): 505-512.
doi: 10.1016/j.tplants.2003.09.004 pmid: 14557048 |
| [8] | Cui BY, Fang SS, Xing YF, et al. Crystallographic analysis of the Arabidopsis thaliana BAG5-calmodulin protein complex[J]. Acta Crystallogr F Struct Biol Commun, 2015, 71(Pt 7): 870-875. |
| [9] | Chen Y, Wang KK, Di J, et al. Mutation of the BAG-1 domain decreases its protective effect against hypoxia/reoxygenation by regulating HSP70 and the PI3K/AKT signalling pathway in SY-SH5Y cells[J]. Brain Res, 2021, 1751: 147192. |
| [10] | Brenner CM, Choudhary M, McC ormick MG, et al. BAG3: Nature's quintessential multi-functional protein functions as a ubiquitous intra-cellular glue[J]. Cells, 2023, 12(6): 937. |
| [11] | Kang CH, Lee JH, Kim YJ, et al. Characterization of AtBAG2 as a novel molecular chaperone[J]. Life-Basel, 2023, 13(3): 687. |
| [12] | Baeken MW, Behl C. On the origin of BAG(3)and its consequences for an expansion of BAG3's role in protein homeostasis[J]. J Cell Biochem, 2022, 123(1): 102-114. |
| [13] | Gupta MK, Randhawa PK, Masternak MM. Role of BAG 5 in protein quality control: Double-edged sword?[J]. Front Aging, 2022, 3: 844168. |
| [14] | Huang HM, Liu CX, Yang C, et al. BAG9 confers thermotolerance by regulating cellular redox homeostasis and the stability of heat shock proteins in Solanum lycopersicum[J]. Antioxidants, 2022, 11(8): 1467. |
| [15] | 杨美玲, 贺丽霞, 于玮玮, 等. 新疆野苹果BAG家族成员分离及BAG7基因的功能分析[J]. 南开大学学报:自然科学版, 2022, 55(6): 1-6. |
| Yang ML, He LX, Yu WW, et al. Identification of BAG and functional analysis of BAG7 of malus sieversii[J]. Acta Sci Nat Univ Nankaiensis, 2022, 55(6): 1-6. | |
| [16] | Arif M, Li ZT, Luo Q, et al. The BAG2 and BAG6 genes are involved in multiple abiotic stress tolerances in Arabidopsis thaliana[J]. Int J Mol Sci, 2021, 22(11): 5856. |
| [17] | Zhang HH, Li YR, Dickman MB, et al. Cytoprotective co-chaperone BcBAG1 is a component for fungal development, virulence, and unfolded protein response(UPR)of Botrytis cinerea[J]. Front Microbiol, 2019, 10: 685. |
| [18] | Li YR, Williams B, Dickman M. Arabidopsis B-cell lymphoma2(Bcl-2)-associated athanogene 7(BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29[J]. New Phytol, 2017, 214(2): 695-705. |
| [19] |
Li LH, Xing YF, Chang D, et al. CaM/BAG5/Hsc70 signaling complex dynamically regulates leaf senescence[J]. Sci Rep, 2016, 6: 31889.
doi: 10.1038/srep31889 pmid: 27539741 |
| [20] | Li YR, Dickman M. Processing of AtBAG6 triggers autophagy and fungal resistance[J]. Plant Signal Behav, 2016, 11(6): e1175699. |
| [21] | Ding HD, Qian L, Jiang HL, et al. Overexpression of a Bcl-2-associated athanogene SlBAG9 negatively regulates high-temperature response in tomato[J]. Int J Biol Macromol, 2022, 194: 695-705. |
| [22] | Yan JQ, He CX, Zhang H. The BAG-family proteins in Arabidopsis thaliana[J]. Plant Sci, 2003, 165(1): 1-7. |
| [23] | Zhou H, Li JY, Liu XY, et al. The divergent roles of the rice bcl-2 associated athanogene(BAG)genes in plant development and environmental responses[J]. Plants, 2021, 10(10): 2169. |
| [24] |
Ge SM, Kang Z, Li Y, et al. Cloning and function analysis of BAG family genes in wheat[J]. Funct Plant Biol, 2016, 43(5): 393-402.
doi: 10.1071/FP15317 pmid: 32480470 |
| [25] | Yeckel G. Characterization of a soybean BAG gene and its potential role in nematode resistance[D]. Columbia: University of Missouri, 2012. |
| [26] | Jiang HL, Ji YR, Sheng JR, et al. Genome-wide identification of the Bcl-2 associated athanogene(BAG)gene family in Solanum lycopersicum and the functional role of SlBAG9 in response to osmotic stress[J]. Antioxidants, 2022, 11(3): 598. |
| [27] | 莫心怡, 周海连, 何丹丹, 等. 甘蔗割手密BAG基因家族的鉴定与表达分析[J]. 基因组学与应用生物学, 2023, 42(12): 1364-1378. |
| Mo XY, Zhou HL, He DD, et al. Identification and expression analysis of BAG gene family in sugarcane saccharum spontaneum[J]. Genom Appl Biol, 2023, 42(12): 1364-1378. | |
| [28] | 张扬, 杜琳, 唐贤丰, 等. 杨树BAG基因的鉴定及表达模式分析[J]. 林业科学, 2019, 55(1): 138-145. |
| Zhang Y, Du L, Tang XF, et al. Identification and expression pattern analyses of Populus BAG genes[J]. Sci Silvae Sin, 2019, 55(1): 138-145. | |
| [29] | Dash A, Ghag SB. Genome-wide in silico characterization and stress induced expression analysis of BcL-2 associated athanogene(BAG)family in Musa spp[J]. Sci Rep, 2022, 12(1): 625. |
| [30] | Lee DW, Kim SJ, Oh YJ, et al. Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins[J]. Mol Plant, 2016, 9(10): 1428-1431. |
| [31] | You QY, Zhai KR, Yang DL, et al. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance[J]. Cell Host Microbe, 2016, 20(6): 758-769. |
| [32] |
Locascio A, Marqués MC, García-Martínez G, et al. BCL2-associated athanogene4 regulates the KAT1 potassium channel and controls stomatal movement[J]. Plant Physiol, 2019, 181(3): 1277-1294.
doi: 10.1104/pp.19.00224 pmid: 31451552 |
| [33] | He MM, Wang Y, Jahan MS, et al. Characterization of SlBAG genes from Solanum lycopersicum and its function in response to dark-induced leaf senescence[J]. Plants, 2021, 10(5): 947. |
| [34] | Irfan M, Kumar P, Ahmad I, et al. Unraveling the role of tomato Bcl-2-associated athanogene(BAG)proteins during abiotic stress response and fruit ripening[J]. Sci Rep, 2021, 11(1): 21734. |
| [35] | Shang KJ, Xiao L, Zhang XP, et al. Tomato chlorosis virus p22 interacts with NbBAG5 to inhibit autophagy and regulate virus infection[J]. Mol Plant Pathol, 2023, 24(5): 425-435. |
| [36] | Jiang HL, Liu XY, Xiao PX, et al. Functional insights of plant bcl-2-associated ahanogene(BAG)proteins: Multi-taskers in diverse cellular signal transduction pathways[J]. Front Plant Sci, 2023, 14: 1136873. |
| [37] |
张开京, 何帅帅, 贾利, 等. 黄瓜DIR家族基因的全基因组鉴定及其表达分析[J]. 中国农业科学, 2023, 56(4): 711-728.
doi: 10.3864/j.issn.0578-1752.2023.04.010 |
| Zhang KJ, He SS, Jia L, et al. Genome-wide identification and expression analysis of DlR gene family in cucumbers[J]. Sci Agric Sin, 2023, 56(4): 711-728. | |
| [38] | 赵振翔, 敖文红, 王新法, 等. 黄瓜DME基因家族的全基因组鉴别及转录分析[J]. 植物生理学报, 2023, 59(1): 209-218. |
| Zhao ZX, Ao WH, Wang XF, et al. Genome-wide identification and transcriptional analysis of DME gene family in cucumber[J]. Plant Physiol J, 2023, 59(1): 209-218. | |
| [39] | 应奥, 程志华, 张小兰, 等. 黄瓜YABBYs家族及CsYAB1a基因功能初探[J]. 中国农业大学学报, 2022, 27(11): 60-78. |
| Ying A, Cheng ZH, Zhang XL, et al. Preliminary study on the function of YABBYs family and CsYAB1a gene in cucumber.(Cucumis sativus L.)[J]. J China Agric Univ, 2022, 27(11): 60-78. | |
| [40] |
周国彦, 银珊珊, 高佳鑫, 等. 黄瓜AHP基因家族的鉴定及其非生物胁迫表达分析[J]. 生物技术通报, 2022, 38(6): 112-119.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1338 |
| Zhou GY, Yin SS, Gao JX, et al. Identification of AHP gene family in cucumis sativus and its expression analysis under abiotic stress[J]. Biotechnol Bull, 2022, 38(6): 112-119. | |
| [41] | Xu MY, Wang YP, Zhang MT, et al. Genome-wide identification of BES1 gene family in six cucurbitaceae species and its expression analysis in Cucurbita moschata[J]. Int J Mol Sci, 2023, 24(3): 2287. |
| [42] | Davoudi M, Chen JF, Lou QF. Genome-wide identification and expression analysis of heat shock protein 70(HSP70)gene family in pumpkin(Cucurbita moschata)rootstock under drought stress suggested the potential role of these chaperones in stress tolerance[J]. Int J Mol Sci, 2022, 23(3): 1918. |
| [43] | Hu YP, Zhang TT, Liu Y, et al. Pumpkin(Cucurbita moschata)HSP20 gene family identification and expression under heat stress[J]. Front Genet, 2021, 12: 753953. |
| [44] | Miao L, Li SZ, Shi AK, et al. Genome-wide analysis of the AINTEGUMENTA-like(AIL)transcription factor gene family in pumpkin(Cucurbita moschata Duch.) and CmoANT1.2 response in graft union healing[J]. Plant Physiol Biochem, 2021, 162: 706-715. |
| [45] | Li Q, Li HB, Huang W, et al. A chromosome-scale genome assembly of cucumber(Cucumis sativus L.)[J]. Gigascience, 2019, 8(6): giz072. |
| [46] |
Sun HH, Wu S, Zhang GY, et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid cucurbita genomes[J]. Mol Plant, 2017, 10(10): 1293-1306.
doi: S1674-2052(17)30266-6 pmid: 28917590 |
| [47] | Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching[J]. Nucleic Acids Res, 2011, 39(Web Server issue): W29-W37. |
| [48] | Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021[J]. Nucleic Acids Res, 2021, 49(D1): D412-D419. |
| [49] | Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020[J]. Nucleic Acids Res, 2021, 49(D1): D458-D460. |
| [50] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [51] | Bailey TL, Williams N, Misleh C, et al. MEME: discovering and analyzing DNA and protein sequence motifs[J]. Nucleic Acids Res, 2006, 34(Web Server issue): W369-W373. |
| [52] |
Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11[J]. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [53] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [54] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity[J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [55] |
Li Z, Zhang ZH, Yan PC, et al. RNA-Seq improves annotation of protein-coding genes in the cucumber genome[J]. BMC Genomics, 2011, 12: 540.
doi: 10.1186/1471-2164-12-540 pmid: 22047402 |
| [56] | Chen CH, Chen XQ, Han J, et al. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses[J]. BMC Plant Biol, 2020, 20(1): 443. |
| [57] | Zhu Y, Yin JL, Liang YF, et al. Transcriptomic dynamics provide an insight into the mechanism for silicon-mediated alleviation of salt stress in cucumber plants[J]. Ecotoxicol Environ Saf, 2019, 174: 245-254. |
| [58] |
Zhou Y, Yang K, Cheng M, et al. Double-faced role of Bcl-2-associated athanogene 7 in plant-Phytophthora interaction[J]. J Exp Bot, 2021, 72(15): 5751-5765.
doi: 10.1093/jxb/erab252 pmid: 34195821 |
| [59] | Hu LZ, Chen JT, Guo JJ, et al. Functional divergence and evolutionary dynamics of BAG gene family in maize(Zea mays)[J]. Int J Agric Biol, 2013, 15(2): 200-206. |
| [60] | UN Food and Agriculture Organization, Corporate Statistical Database(FAOSTAT). Production of cucumbers and gherkins in 2020[EB/OL]. https://www.fao.org/faostat/zh/#data/.2020-03-30/2023-12-18. |
| [61] | Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice(Oryza sativa)[J]. Genomics, 2006, 88(3): 360-371. |
| [62] |
Zhang SB, C hen C, Li L, et al. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family[J]. Plant Physiol, 2005, 139(3): 1107-1124.
pmid: 16286450 |
| [63] |
Nollen EA, Brunsting JF, Song J, et al. Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity[J]. Mol Cell Biol, 2000, 20(3): 1083-1088.
doi: 10.1128/MCB.20.3.1083-1088.2000 pmid: 10629065 |
| [64] |
Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators[J]. J Biol Chem, 1999, 274(2): 781-786.
doi: 10.1074/jbc.274.2.781 pmid: 9873016 |
| [65] |
Takayama S, Bimston DN, Matsuzawa S, et al. BAG-1 modulates the chaperone activity of Hsp70/Hsc70[J]. EMBO J, 1997, 16(16): 4887-4896.
doi: 10.1093/emboj/16.16.4887 pmid: 9305631 |
| [66] | Nawkar GM, Maibam P, Park JH, et al. In silico study on Arabidopsis BAG gene expression in response to environmental stresses[J]. Protoplasma, 2017, 254(1): 409-421. |
| [67] | Wang J, Nan N, Li N, et al. A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress[J]. Plant Cell, 2020, 32(11): 3535-3558. |
| [68] |
Wang JY, Yeckel G, Kandoth PK, et al. Targeted suppression of soybean BAG6-induced cell death in yeast by soybean cyst nematode effectors[J]. Mol Plant Pathol, 2020, 21(9): 1227-1239.
doi: 10.1111/mpp.12970 pmid: 32686295 |
| [1] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [2] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [3] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [4] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [5] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [6] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [7] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [8] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [9] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [10] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [11] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [12] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [13] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [14] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [15] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||