








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 167-178.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1092
Previous Articles Next Articles
HOU Ya-qiong1( ), LANG Hong-shan1, WEN Meng-meng1, GU Yi-yun2, ZHU Run-jie2, TANG Xiao-li1(
), LANG Hong-shan1, WEN Meng-meng1, GU Yi-yun2, ZHU Run-jie2, TANG Xiao-li1( )
)
Received:2023-11-20
Online:2024-05-26
Published:2024-06-13
Contact:
TANG Xiao-li
E-mail:houyaqiong2023@163.com;tangxiaoli.1@163.com
HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit[J]. Biotechnology Bulletin, 2024, 40(5): 167-178.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| AcHSP20-2 | GATCTATCGCACCGTGTC | AACCTTTATGTCGTCCTTG |
| AcHSP20-5 | CTTCCAGGAGGTGTATCGT | CGTCTTCTTTGGCATTGTA |
| AcHSP20-9 | TCTCACCTTTGGGTTTATTG | CAGGGAACGTCATGGAA |
| AcHSP20-12 | CGGGAGGAGGAGAAAGAA | CAAACAGCAGACACGGC |
| AcHSP20-19 | AGAAGAATGGAGGTGATGAT | GAGATGGAGCACAAGAGTTT |
| AcActin | TGCATGAGCGATCAAGTTTCAAG | TGTCCCATGTCTGGTTGATGACT |
Table 1 Primers used in the expression analysis of AcHSP20s genes
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| AcHSP20-2 | GATCTATCGCACCGTGTC | AACCTTTATGTCGTCCTTG |
| AcHSP20-5 | CTTCCAGGAGGTGTATCGT | CGTCTTCTTTGGCATTGTA |
| AcHSP20-9 | TCTCACCTTTGGGTTTATTG | CAGGGAACGTCATGGAA |
| AcHSP20-12 | CGGGAGGAGGAGAAAGAA | CAAACAGCAGACACGGC |
| AcHSP20-19 | AGAAGAATGGAGGTGATGAT | GAGATGGAGCACAAGAGTTT |
| AcActin | TGCATGAGCGATCAAGTTTCAAG | TGTCCCATGTCTGGTTGATGACT |
| 基因 Gene | 基因组登录号 Gene ID | 等电点 pI | 分子量 Molecular weight/kD | 氨基酸数Number of amino acids | 染色体定位及基因方向Chromosome localization and gene orientation | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|
| AcHSP20-1 | Ach00g218421.2 | 6.57 | 10.16 | 88 | Chr00: 54153215.. 54154904(-) | 细胞质Cytoplasmic |
| AcHSP20-2 | Ach00g239181 | 7.86 | 16.14 | 143 | Chr00: 50739806.. 50740562(-) | 过氧化物酶体Peroxisomal |
| AcHSP20-3 | Ach00g329621 | 5.59 | 23.28 | 208 | Chr00: 90841963.. 90843153(-) | 叶绿体Chloroplast |
| AcHSP20-4 | Ach00g479741.2 | 10.32 | 9.93 | 86 | Chr00: 135211617.. 135211877(+) | 细胞质Cytoplasmic |
| AcHSP20-5 | Ach00g479761.2 | 9.14 | 40.18 | 371 | Chr00: 145805825.. 145808080(-) | 高尔基体Golgi apparatus |
| AcHSP20-6 | Ach02g036831 | 7.8 | 16.56 | 152 | Chr02: 4423490.. 4423948(-) | 细胞质Cytoplasmic |
| AcHSP20-7 | Ach02g046741.2 | 6.21 | 25.55 | 225 | Chr02: 516872.. 518080(-) | 叶绿体Chloroplast |
| AcHSP20-8 | Ach02g223291 | 8.86 | 17.61 | 153 | Chr02: 4090770.. 4091231(-) | 细胞质Cytoplasmic |
| AcHSP20-9 | Ach03g045661 | 7.63 | 25.69 | 232 | Chr03: 6797048.. 6798489(-) | 叶绿体Chloroplast |
| AcHSP20-10 | Ach03g451751.2 | 9.15 | 11.12 | 96 | Chr03: 1045510.. 1045800(-) | 叶绿体Chloroplast |
| AcHSP20-11 | Ach04g161211.2 | 6.45 | 24.77 | 215 | Chr04: 6179073.. 6180486(-) | 细胞质Cytoplasmic |
| AcHSP20-12 | Ach06g418911.2 | 6.32 | 16.60 | 152 | Chr06: 2925504.. 2925962(-) | 细胞质Cytoplasmic |
| AcHSP20-13 | Ach10g027441.2 | 5.4 | 20.33 | 181 | Chr10: 4989172.. 4989717(-) | 细胞质Cytoplasmic |
| AcHSP20-14 | Ach10g027451 | 7.77 | 25.72 | 228 | Chr10: 4973842.. 4974528(-) | 细胞质Cytoplasmic |
| AcHSP20-15 | Ach10g027461.2 | 6.34 | 24.95 | 221 | Chr10: 4984511.. 4985176(+) | 叶绿体Chloroplast |
| AcHSP20-16 | Ach10g143161 | 9.3 | 25.78 | 228 | Chr10: 11814664.. 11815350(+) | 细胞质Cytoplasmic |
| AcHSP20-17 | Ach11g263431.2 | 5.05 | 21.72 | 196 | Chr11: 8776984.. 8777574(-) | 细胞质Cytoplasmic |
| AcHSP20-18 | Ach11g425231.2 | 4.43 | 10.59 | 90 | Chr11: 12263315.. 12263587(-) | 细胞质Cytoplasmic |
| AcHSP20-19 | Ach12g058351.2 | 8.69 | 17.21 | 152 | Chr12: 6955029.. 6956940(+) | 细胞核Nucleus |
| AcHSP20-20 | Ach13g308811 | 5.94 | 33.07 | 297 | Chr13: 1116598.. 1117580(+) | 叶绿体Chloroplast |
| AcHSP20-21 | Ach13g350901 | 5.68 | 21.90 | 195 | Chr13: 14397259.. 14398212(+) | 叶绿体Chloroplast |
| AcHSP20-22 | Ach13g383281 | 6.31 | 16.61 | 152 | Chr13: 11985660.. 11986118(-) | 细胞质Cytoplasmic |
| AcHSP20-23 | Ach13g472651.2 | 5.94 | 33.07 | 297 | Chr13: 1497079.. 1498061(-) | 叶绿体Chloroplast |
| AcHSP20-24 | Ach14g205801.2 | 6.87 | 24.06 | 216 | Chr14: 9422059.. 9423375(+) | 叶绿体Chloroplast |
| AcHSP20-25 | Ach14g205811.2 | 8.86 | 36.76 | 329 | Chr14: 9424474.. 9428179(+) | 叶绿体Chloroplast |
| AcHSP20-26 | Ach15g376651 | 5.67 | 22.90 | 202 | Chr15: 4758959.. 4759567(-) | 细胞质Cytoplasmic |
| AcHSP20-27 | Ach16g309411.2 | 7.64 | 26.14 | 231 | Chr16: 8508810.. 8510240(-) | 叶绿体Chloroplast |
| AcHSP20-28 | Ach16g331111 | 8.83 | 17.36 | 153 | Chr16: 10507526.. 10508507(+) | 细胞质Cytoplasmic |
| AcHSP20-29 | Ach18g165371 | 4.99 | 22.60 | 198 | Chr18: 8741760.. 8742825(+) | 细胞质Cytoplasmic |
| AcHSP20-30 | Ach19g138461.2 | 5.08 | 13.25 | 114 | Chr19: 7272801.. 7273145(+) | 细胞质Cytoplasmic |
| AcHSP20-31 | Ach22g015991.2 | 7.61 | 26.63 | 241 | Chr22: 8846412.. 8851047(-) | 叶绿体Chloroplast |
| AcHSP20-32 | Ach23g121711.2 | 5.37 | 22.34 | 199 | Chr23: 4320454.. 4321535(-) | 细胞质Cytoplasmic |
| AcHSP20-33 | Ach23g411051.2 | 9.11 | 33.06 | 300 | Chr23: 19087591.. 19088582(+) | 叶绿体Chloroplast |
| AcHSP20-34 | Ach29g196461.2 | 4.59 | 9.84 | 85 | Chr29: 3710817.. 3711074(+) | 叶绿体Chloroplast |
Table 2 Physicochemical properties of AcHSP20s proteins in kiwifruit
| 基因 Gene | 基因组登录号 Gene ID | 等电点 pI | 分子量 Molecular weight/kD | 氨基酸数Number of amino acids | 染色体定位及基因方向Chromosome localization and gene orientation | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|
| AcHSP20-1 | Ach00g218421.2 | 6.57 | 10.16 | 88 | Chr00: 54153215.. 54154904(-) | 细胞质Cytoplasmic |
| AcHSP20-2 | Ach00g239181 | 7.86 | 16.14 | 143 | Chr00: 50739806.. 50740562(-) | 过氧化物酶体Peroxisomal |
| AcHSP20-3 | Ach00g329621 | 5.59 | 23.28 | 208 | Chr00: 90841963.. 90843153(-) | 叶绿体Chloroplast |
| AcHSP20-4 | Ach00g479741.2 | 10.32 | 9.93 | 86 | Chr00: 135211617.. 135211877(+) | 细胞质Cytoplasmic |
| AcHSP20-5 | Ach00g479761.2 | 9.14 | 40.18 | 371 | Chr00: 145805825.. 145808080(-) | 高尔基体Golgi apparatus |
| AcHSP20-6 | Ach02g036831 | 7.8 | 16.56 | 152 | Chr02: 4423490.. 4423948(-) | 细胞质Cytoplasmic |
| AcHSP20-7 | Ach02g046741.2 | 6.21 | 25.55 | 225 | Chr02: 516872.. 518080(-) | 叶绿体Chloroplast |
| AcHSP20-8 | Ach02g223291 | 8.86 | 17.61 | 153 | Chr02: 4090770.. 4091231(-) | 细胞质Cytoplasmic |
| AcHSP20-9 | Ach03g045661 | 7.63 | 25.69 | 232 | Chr03: 6797048.. 6798489(-) | 叶绿体Chloroplast |
| AcHSP20-10 | Ach03g451751.2 | 9.15 | 11.12 | 96 | Chr03: 1045510.. 1045800(-) | 叶绿体Chloroplast |
| AcHSP20-11 | Ach04g161211.2 | 6.45 | 24.77 | 215 | Chr04: 6179073.. 6180486(-) | 细胞质Cytoplasmic |
| AcHSP20-12 | Ach06g418911.2 | 6.32 | 16.60 | 152 | Chr06: 2925504.. 2925962(-) | 细胞质Cytoplasmic |
| AcHSP20-13 | Ach10g027441.2 | 5.4 | 20.33 | 181 | Chr10: 4989172.. 4989717(-) | 细胞质Cytoplasmic |
| AcHSP20-14 | Ach10g027451 | 7.77 | 25.72 | 228 | Chr10: 4973842.. 4974528(-) | 细胞质Cytoplasmic |
| AcHSP20-15 | Ach10g027461.2 | 6.34 | 24.95 | 221 | Chr10: 4984511.. 4985176(+) | 叶绿体Chloroplast |
| AcHSP20-16 | Ach10g143161 | 9.3 | 25.78 | 228 | Chr10: 11814664.. 11815350(+) | 细胞质Cytoplasmic |
| AcHSP20-17 | Ach11g263431.2 | 5.05 | 21.72 | 196 | Chr11: 8776984.. 8777574(-) | 细胞质Cytoplasmic |
| AcHSP20-18 | Ach11g425231.2 | 4.43 | 10.59 | 90 | Chr11: 12263315.. 12263587(-) | 细胞质Cytoplasmic |
| AcHSP20-19 | Ach12g058351.2 | 8.69 | 17.21 | 152 | Chr12: 6955029.. 6956940(+) | 细胞核Nucleus |
| AcHSP20-20 | Ach13g308811 | 5.94 | 33.07 | 297 | Chr13: 1116598.. 1117580(+) | 叶绿体Chloroplast |
| AcHSP20-21 | Ach13g350901 | 5.68 | 21.90 | 195 | Chr13: 14397259.. 14398212(+) | 叶绿体Chloroplast |
| AcHSP20-22 | Ach13g383281 | 6.31 | 16.61 | 152 | Chr13: 11985660.. 11986118(-) | 细胞质Cytoplasmic |
| AcHSP20-23 | Ach13g472651.2 | 5.94 | 33.07 | 297 | Chr13: 1497079.. 1498061(-) | 叶绿体Chloroplast |
| AcHSP20-24 | Ach14g205801.2 | 6.87 | 24.06 | 216 | Chr14: 9422059.. 9423375(+) | 叶绿体Chloroplast |
| AcHSP20-25 | Ach14g205811.2 | 8.86 | 36.76 | 329 | Chr14: 9424474.. 9428179(+) | 叶绿体Chloroplast |
| AcHSP20-26 | Ach15g376651 | 5.67 | 22.90 | 202 | Chr15: 4758959.. 4759567(-) | 细胞质Cytoplasmic |
| AcHSP20-27 | Ach16g309411.2 | 7.64 | 26.14 | 231 | Chr16: 8508810.. 8510240(-) | 叶绿体Chloroplast |
| AcHSP20-28 | Ach16g331111 | 8.83 | 17.36 | 153 | Chr16: 10507526.. 10508507(+) | 细胞质Cytoplasmic |
| AcHSP20-29 | Ach18g165371 | 4.99 | 22.60 | 198 | Chr18: 8741760.. 8742825(+) | 细胞质Cytoplasmic |
| AcHSP20-30 | Ach19g138461.2 | 5.08 | 13.25 | 114 | Chr19: 7272801.. 7273145(+) | 细胞质Cytoplasmic |
| AcHSP20-31 | Ach22g015991.2 | 7.61 | 26.63 | 241 | Chr22: 8846412.. 8851047(-) | 叶绿体Chloroplast |
| AcHSP20-32 | Ach23g121711.2 | 5.37 | 22.34 | 199 | Chr23: 4320454.. 4321535(-) | 细胞质Cytoplasmic |
| AcHSP20-33 | Ach23g411051.2 | 9.11 | 33.06 | 300 | Chr23: 19087591.. 19088582(+) | 叶绿体Chloroplast |
| AcHSP20-34 | Ach29g196461.2 | 4.59 | 9.84 | 85 | Chr29: 3710817.. 3711074(+) | 叶绿体Chloroplast |

Fig. 2 Chromosomal localization and synteny analysis of AcHSP20s A: Chromosomal localizations of 34 AcHSP20s in kiwifruit. B: Synteny analysis of 34 AcHSP20s in kiwifruit. C: Synteny analysis between 34 AcHSP20s in kiwifruit and 19 AtHSP20s in Arabidopsis
| 基因对Gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| AcHSP20-7/AcHSP20-10 | 0.231 715 656 | 0.423 821 440 | 0.546 729 433 |
| AcHSP20-7/AcHSP20-9 | 0.209 150 940 | 0.704 599 321 | 0.296 836 704 |
| AcHSP20-10/AcHSP20-9 | 0.322 818 372 | 0.919 187 813 | 0.351 199 578 |
| AcHSP20-18/AcHSP20-27 | 0.102 831 910 | 0.278 905 623 | 0.368 697 875 |
| AcHSP20-20/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-23/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-21/AcHSP20-34 | 0.092 066 167 | 0.397 378 202 | 0.231 683 988 |
| AcHSP20-30/AcHSP20-32 | 0.239 953 460 | 0.526 314 021 | 0.455 913 107 |
Table 3 Ka/Ks value of AcHSP20s
| 基因对Gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| AcHSP20-7/AcHSP20-10 | 0.231 715 656 | 0.423 821 440 | 0.546 729 433 |
| AcHSP20-7/AcHSP20-9 | 0.209 150 940 | 0.704 599 321 | 0.296 836 704 |
| AcHSP20-10/AcHSP20-9 | 0.322 818 372 | 0.919 187 813 | 0.351 199 578 |
| AcHSP20-18/AcHSP20-27 | 0.102 831 910 | 0.278 905 623 | 0.368 697 875 |
| AcHSP20-20/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-23/AcHSP20-33 | 0.104 764 648 | 0.138 864 084 | 0.754 440 205 |
| AcHSP20-21/AcHSP20-34 | 0.092 066 167 | 0.397 378 202 | 0.231 683 988 |
| AcHSP20-30/AcHSP20-32 | 0.239 953 460 | 0.526 314 021 | 0.455 913 107 |

Fig. 4 Investigation of cis-acting elements in the promoters of AcHSP20s A: The locations of cis-acting elements of 34 AcHSP20s in kiwifruit. B: Numbers of 34 AcHSP20s cis-acting elements in kiwifruit
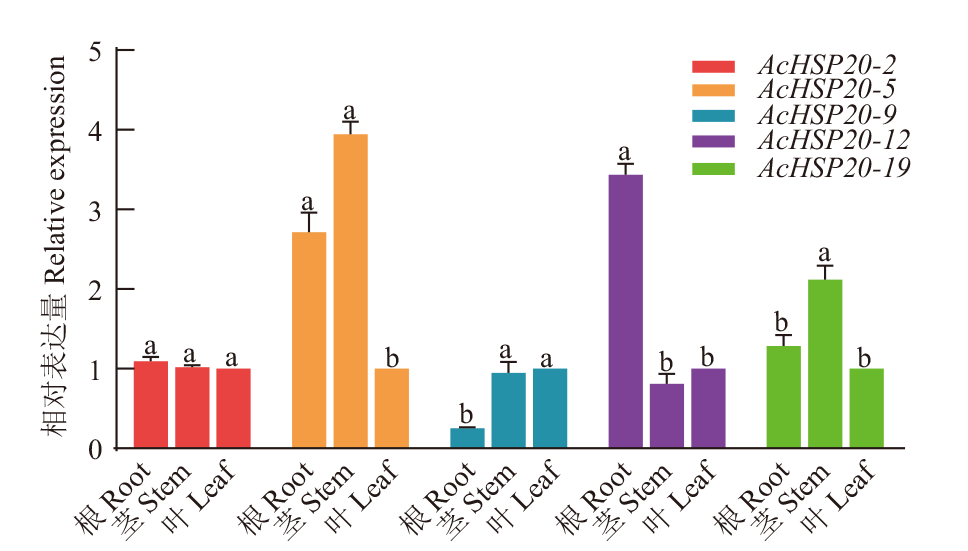
Fig. 5 Expression profiles of AcHSP20s in kiwifruit There are 3 biological and technical replicates, the standard error is on the bar graph, and lowercase letters indicate the difference among different samples. The same below
| [1] |
Herman DJ, Knowles LO, Knowles NR. Heat stress affects carbohydrate metabolism during cold-induced sweetening of potato(Solanum tuberosum L.)[J]. Planta, 2017, 245(3): 563-582.
doi: 10.1007/s00425-016-2626-z pmid: 27904974 |
| [2] |
Giorno F, Wolters-Arts M, Grillo S, et al. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers[J]. J Exp Bot, 2010, 61(2): 453-462.
doi: 10.1093/jxb/erp316 pmid: 19854799 |
| [3] |
Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants[J]. Curr Opin Plant Biol, 2011, 14(3): 290-295.
doi: 10.1016/j.pbi.2011.02.001 pmid: 21377404 |
| [4] |
Wang N, Guo TL, Sun X, et al. Functions of two Malus hupehensis(Pamp.) Rehd. YTPs(MhYTP1 and MhYTP2)in biotic- and abiotic-stress responses[J]. Plant Sci, 2017, 261: 18-27.
doi: S0168-9452(17)30238-8 pmid: 28554690 |
| [5] | Suzuki N. Temperature stress and responses in plants[J]. Int J Mol Sci, 2019, 20(8): 2001. |
| [6] |
Yang GD, Yu ZP, Gao L, et al. SnRK2s at the crossroads of growth and stress responses[J]. Trends Plant Sci, 2019, 24(8): 672-676.
doi: S1360-1385(19)30139-6 pmid: 31255544 |
| [7] |
Tang XL, Mu XM, Shao HB, et al. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology[J]. Crit Rev Biotechnol, 2015, 35(4): 425-437.
doi: 10.3109/07388551.2014.889080 pmid: 24738851 |
| [8] | Tang XL, Shao HB, Jiang FD, et al. Molecular cloning and functional analyses of the salt-responsive gene KvHSP70 from Kosteletzkya virginica[J]. Land Degrad Dev, 2020, 31(6): 773-782. |
| [9] | Wu JT, Gao T, Hu JN, et al. Research advances in function and regulation mechanisms of plant small heat shock proteins(sHSPs)under environmental stresses[J]. Sci Total Environ, 2022, 825: 154054. |
| [10] |
Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance[J]. Plant Biotechnol J, 2017, 15(4): 405-414.
doi: 10.1111/pbi.12659 pmid: 27860233 |
| [11] | Zhang KM, Ezemaduka AN, Wang Z, et al. A novel mechanism for small heat shock proteins to function as molecular chaperones[J]. Sci Rep, 2015, 5(5): 8811-8819. |
| [12] | Haq S U, Khan A, Ali M, et al. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses[J]. Int J Mol Sci, 2019, 20(21): 5321. |
| [13] | Tian C, Zhang ZY, Huang Y, et al. Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco[J]. Plant Physiol Biochem, 2022, 177: 1-9. |
| [14] | Feng XH, Zhang HX, Ali M, et al. A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper(Capsicum annuum L.)[J]. Plant Physiol Biochem, 2019, 142(142): 151-162. |
| [15] | Waters ER. The evolution, function, structure and expression of the plant sHSPs[J]. J Exp Bot, 2013, 64(2): 391-403. |
| [16] | Waters ER, Vierling E. Plant small heat shock proteins-evolutionary and functional diversity[J]. New Phytol, 2020, 227(1): 24-37. |
| [17] |
Haslbeck M, Franzmann T, Weinfurtner D, et al. Some like it hot: the structure and function of small heat-shock proteins[J]. Nat Struct Mol Biol, 2005, 12(10): 842-846.
doi: 10.1038/nsmb993 pmid: 16205709 |
| [18] | El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019[J]. Nucleic Acids Res, 2019, 47(D1): D427-D432. |
| [19] |
Ma W, Guan XY, Li J, et al. Mitochondrial small heat shock protein mediates seed germination via thermal sensing[J]. Proc Natl Acad Sci USA, 2019, 116(10): 4716-4721.
doi: 10.1073/pnas.1815790116 pmid: 30765516 |
| [20] |
Rutsdottir G, Härmark J, Weide Y, et al. Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity[J]. J Biol Chem, 2017, 292(19): 8103-8121.
doi: 10.1074/jbc.M116.766816 pmid: 28325834 |
| [21] |
Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation[J]. EMBO J, 2008, 27(2): 328-335.
doi: 10.1038/sj.emboj.7601970 pmid: 18216875 |
| [22] | Yang ML, Zhang YX, Zhang HH, et al. Identification of MsHsp20 gene family in malus sieversii and functional characterization of MsHsp16.9 in heat tolerance[J]. Front Plant Sci, 2017, 8: 1761-1778. |
| [23] | Sewelam N, Kazan K, Hüdig M, et al. The AtHSP17.4C1 gene expression is mediated by diverse signals that link biotic and abiotic stress factors with ROS and can be a useful molecular marker for oxidative stress[J]. Int J Mol Sci, 2019, 20(13): 3201-3218. |
| [24] | Mu CJ, Zhang SJ, Yu GZ, et al. Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses[J]. PLoS One, 2013, 8(12): e82264-e82273. |
| [25] | Zhang L, Gao YK, Pan HT, et al. Cloning and characterisation of a Primula heat shock protein gene, PfHSP17.1, which confers heat, salt and drought tolerance in transgenic Arabidopsis thaliana[J]. Acta Physiol Plant, 2013, 35(11): 3191-3200. |
| [26] | Sun XB, Sun CY, Li ZG, et al. AsHSP17, a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abiotic stress[J]. Plant Cell Environ, 2016, 39(6): 1320-1337. |
| [27] | Zhang N, Zhao HY, Shi JW, et al. Functional characterization of class I SlHSP17.7 gene responsible for tomato cold-stress tolerance[J]. Plant Sci, 2020, 298: 110568. |
| [28] | Do VG, Lee Y, Kweon H, et al. Light induces carotenoid biosynthesis-related gene expression, accumulation of pigment content, and expression of the small heat shock protein in apple fruit[J]. Int J Mol Sci, 2022, 23(11): 6153. |
| [29] |
Kaur H, Petla BP, Kamble NU, et al. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress[J]. Front Plant Sci, 2015, 6: 713.
doi: 10.3389/fpls.2015.00713 pmid: 26442027 |
| [30] |
Kuang J, Liu JZ, Mei J, et al. A class II small heat shock protein OsHsp18.0 plays positive roles in both biotic and abiotic defense responses in rice[J]. Sci Rep, 2017, 7(1): 11333.
doi: 10.1038/s41598-017-11882-x pmid: 28900229 |
| [31] |
Abassi S, Wang H, Ponmani T, et al. Small heat shock protein genes of the green algae Closterium ehrenbergii: Cloning and differential expression under heat and heavy metal stresses[J]. Environ Toxicol, 2019, 34(9): 1013-1024.
doi: 10.1002/tox.22772 pmid: 31095847 |
| [32] | Li ZY, Long RC, Zhang TJ, et al. Molecular cloning and characterization of the MsHSP17.7 gene from Medicago sativa L.[J]. Mol Biol Rep, 2016, 43(8): 815-826. |
| [33] | Li XW, Li JQ, Djaja DS. New synonyms in Actinidiaceae from China[J]. J Syst Evol, 2007, 45(5): 633-660. |
| [34] | 钟彩虹, 黄文俊, 李大卫, 等. 世界猕猴桃产业发展及鲜果贸易动态分析[J]. 中国果树, 2021, 7(7): 101-108. |
| Zhong CH, Huang WJ, Li DW, et al. Dynamic analysis of global kiwifruit industry development and fresh fruit trade[J]. China Fruits, 2021, 7(7): 101-108. | |
| [35] | Hua YG, Liu Q, Zhai YF, et al. Genome-wide analysis of the HSP20 gene family and its response to heat and drought stress in Coix(Coix lacryma-jobi L.)[J]. BMC Genomics, 2023, 24(1): 478-494. |
| [36] | Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains(Acd proteins)[J]. Cell Stress Chaperones, 2001, 6(3): 225-237. |
| [37] |
Ouyang YD, Chen JJ, Xie WB, et al. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice[J]. Plant Mol Biol, 2009, 70(3): 341-357.
doi: 10.1007/s11103-009-9477-y pmid: 19277876 |
| [38] |
Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms[J]. Mol Biol Evol, 2001, 18(9): 1611-1630.
doi: 10.1093/oxfordjournals.molbev.a003951 pmid: 11504843 |
| [39] | Huang LJ, Cheng GX, Khan A, et al. CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance[J]. Protoplasma, 2019, 256(1): 39-51. |
| [40] | He YJ, Yao YX, Li LL, et al. A heat-shock 20 protein isolated from watermelon(ClHSP22.8)negatively regulates the response of Arabidopsis to salt stress via multiple signaling pathways[J]. PeerJ, 2021, 9: e10524. |
| [1] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [2] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [3] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [4] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [5] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [6] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [7] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [8] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| [9] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [10] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [11] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [12] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [13] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [14] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [15] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||