








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 238-250.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0044
Previous Articles Next Articles
CHANG Xue-rui( ), WANG Tian-tian, WANG Jing(
), WANG Tian-tian, WANG Jing( )
)
Received:2024-01-12
Online:2024-06-26
Published:2024-05-23
Contact:
WANG Jing
E-mail:C15835567493@163.com;wangjing315@sxau.edu.cn
CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.)[J]. Biotechnology Bulletin, 2024, 40(6): 238-250.
| 基因名称 Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CaUBC3 | AGCGGAGGTAGAGGAAGAGG | GCCACCCTCATAAGGTGTTC |
| CaUBC8 | GCTGGTGGTGTTTTCCAAGT | GCTTCGAGCCATTGATTCAT |
| CaUBC18 | TGCAGGTGGTGTTTTTCAAG | CGAGCCACCGATTCATATTT |
| CaUBC21 | ATAGCCCCTATGCAGGAGGT | CAAGTGGATCGTCTGGGTTT |
| CaUBC23 | CCCAATAAGCCACCAACAGT | ACTGAACATCCGAGCTGCTT |
| CaUBC38 | GCGGAAAGTCCTTATCATGG | GATCAACAGGACCTGCCATT |
| β-Actin | CCACCTCTTCACTCTCTGCTCT | ACTAGGAAAAACAGCCCTTGGT |
Table 1 Primers required for RT-qPCR for E2 family gene expression analysis
| 基因名称 Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CaUBC3 | AGCGGAGGTAGAGGAAGAGG | GCCACCCTCATAAGGTGTTC |
| CaUBC8 | GCTGGTGGTGTTTTCCAAGT | GCTTCGAGCCATTGATTCAT |
| CaUBC18 | TGCAGGTGGTGTTTTTCAAG | CGAGCCACCGATTCATATTT |
| CaUBC21 | ATAGCCCCTATGCAGGAGGT | CAAGTGGATCGTCTGGGTTT |
| CaUBC23 | CCCAATAAGCCACCAACAGT | ACTGAACATCCGAGCTGCTT |
| CaUBC38 | GCGGAAAGTCCTTATCATGG | GATCAACAGGACCTGCCATT |
| β-Actin | CCACCTCTTCACTCTCTGCTCT | ACTAGGAAAAACAGCCCTTGGT |
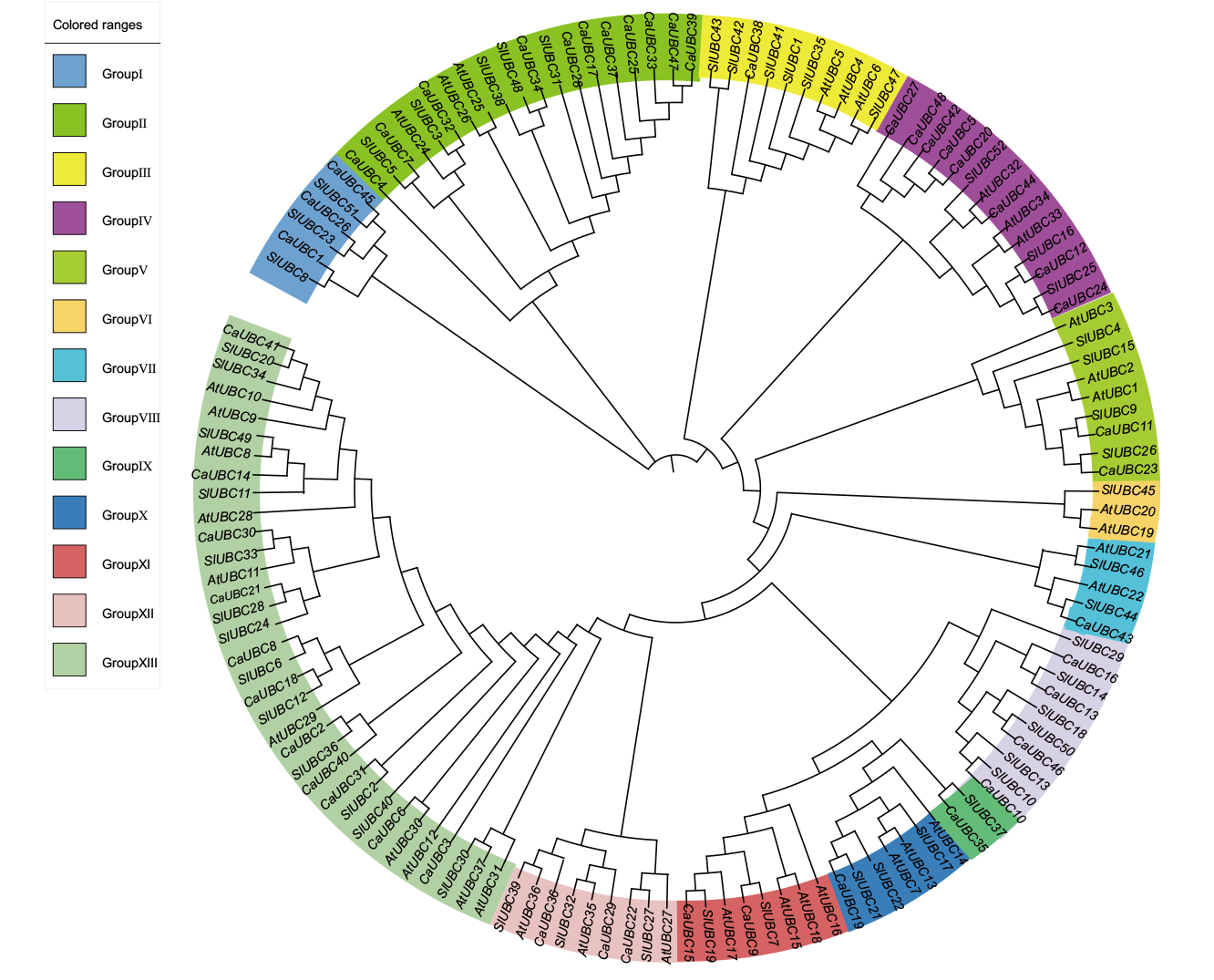
Fig. 2 Phylogenetic tree of UBC protein in C. annuum L.(48),A. thaliana L.(36)and S. lycopersicum L.(52) At: Arabidopsis thaliana L.; Sl: Solanum lycopersicum L.; Ca: Capsicum annuum L.

Fig. 5 Analysis of the protein interaction network of the E2 family The nodes in the protein interaction network indicate all proteins produced by a protein-coding locus, and the nodes of different colors indicate different degrees of interaction, and the intensity of interaction increases with the deepening of the color of the lines between nodes, and the thinner lines indicate that the interaction between proteins is weaker

Fig. 7 Expression profiles of CaUBC genes in different organs F1-F9: After the appearance of flower buds. G1-G11: Fruits at 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, and 60 d after pollination. ST1 and ST2: Fruits at 10 and 15 d after pollination. S3-S11: Fruits at 20, 25, 30, 35, 40, 45, 50, 55, and 60 d after pollination. T3-T11: Fruits at 20, 25, 30, 35, 40, 45, 50, 55 and 60 d after pollination. L1-L9: 2, 5, 10, 15, 20, 25, 30, 40, 50 and 60 d after the emergence of new leaves. The same below

Fig. 8 Relative expressions of eight representative genes of E2 gene family in pepper A: Relative expression of CaUBC38 during fruit development; relative expression of CaUBC8 during flower development; relative expression of CaUBC3/18 under heat stress.B: CaUBC8/21/23/38 under heat stress
| [1] | Parisi M, Alioto D, Tripodi P. Overview of biotic stresses in pepper(Capsicum spp.): sources of genetic resistance, molecular breeding and genomics[J]. Int J Mol Sci, 2020, 21(7): 2587. |
| [2] | Gao CL, Mumtaz MA, Zhou Y, et al. Integrated transcriptomic and metabolomic analyses of cold-tolerant and cold-sensitive pepper species reveal key genes and essential metabolic pathways involved in response to cold stress[J]. Int J Mol Sci, 2022, 23(12): 6683. |
| [3] |
宁约瑟, 王国梁, 谢旗. 泛素连接酶E3介导的植物干旱胁迫反应[J]. 植物学报, 2011, 46(6): 606-616.
doi: 10.3724/SP.J.1259.2011.00606 |
| Ning YS, Wang GL, Xie Q. E3 ubiquitin ligase-mediated drought responses in plants[J]. Chin Bull Bot, 2011, 46(6): 606-616. | |
| [4] |
张蓓, 任福森, 赵洋, 等. 辣椒响应热胁迫机制的研究进展[J]. 生物技术通报, 2023, 39(7): 37-47.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0132 |
|
Zhang B, Ren FS, Zhao Y, et al. Advances in the mechanism of pepper in the response to heat stress[J]. Biotechnol Bull, 2023, 39(7): 37-47.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0132 |
|
| [5] |
Guo M, Yin YX, Ji JJ, et al. Cloning and expression analysis of heat-shock transcription factor gene CaHsfA2 from pepper(Capsicum annuum L.)[J]. Genet Mol Res, 2014, 13(1): 1865-1875.
doi: 10.4238/2014.March.17.14 pmid: 24668674 |
| [6] |
Ernst A, Avvakumov G, Tong JF, et al. A strategy for modulation of enzymes in the ubiquitin system[J]. Science, 2013, 339(6119): 590-595.
doi: 10.1126/science.1230161 pmid: 23287719 |
| [7] |
Sadanandom A, Bailey M, Ewan R, et al. The ubiquitin-proteasome system: central modifier of plant signalling[J]. New Phytol, 2012, 196(1): 13-28.
doi: 10.1111/j.1469-8137.2012.04266.x pmid: 22897362 |
| [8] |
Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling[J]. Science, 2007, 315(5809): 201-205.
doi: 10.1126/science.1127085 pmid: 17218518 |
| [9] | Kraft E, Stone SL, Ma LG, et al. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis[J]. Plant Physiol, 2005, 139(4): 1597-1611. |
| [10] | Stone SL, Hauksdóttir H, Troy A, et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis[J]. Plant Physiol, 2005, 137(1): 13-30. |
| [11] |
俞沁含, 焦淑珍, 吴楠, 等. 葡萄E3泛素酶HOS1基因克隆、表达及抗血清制备[J]. 园艺学报, 2021, 48(6): 1173-1182.
doi: 10.16420/j.issn.0513-353x.2020-0557 |
| Yu QH, Jiao SZ, Wu N, et al. Molecular cloning, expression and polyclonal antibody preparation of E3 ubiquitin ligase gene HOS1 from Vitis vinifera[J]. Acta Hortic Sin, 2021, 48(6): 1173-1182. | |
| [12] | Liu WG, Tang X, Qi XH, et al. The ubiquitin conjugating enzyme: an important ubiquitin transfer platform in ubiquitin-proteasome system[J]. Int J Mol Sci, 2020, 21(8): 2894. |
| [13] | Bae H, Kim WT. The N-terminal tetra-peptide(IPDE)short extension of the U-box motif in rice SPL11 E3 is essential for the interaction with E2 and ubiquitin-ligase activity[J]. Biochem Biophys Res Commun, 2013, 433(2): 266-271. |
| [14] | Bae H, Kim WT. Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases[J]. Biochem Biophys Res Commun, 2014, 444(4): 575-580. |
| [15] | Ye YH, Rape M. Building ubiquitin chains: E2 enzymes at work[J]. Nat Rev Mol Cell Biol, 2009, 10(11): 755-764. |
| [16] |
王春萍, 张世才, 李怡斐, 等. 辣椒泛素结合酶变体类似蛋白基因克隆及其与氮吸收利用的相关性分析[J]. 核农学报, 2023, 37(3): 442-448.
doi: 10.11869/j.issn.1000-8551.2023.03.0442 |
|
Wang CP, Zhang SC, Li YF, et al. Cloning of E2 variant like protein gene in pepper and its correlation with nitrogen absorption and utilization[J]. J Nucl Agric Sci, 2023, 37(3): 442-448.
doi: 10.11869/j.issn.1000-8551.2023.03.0442 |
|
| [17] | 顾星. 人类泛素结合酶UBE2T、UBE2W和UBE2Z的功能研究[D]. 上海: 复旦大学, 2008. |
| Gu X. Functional studies of human ubiquitin-conjugating enzymes UBE2T, UBE2W, and UBE2Z[D]. Shanghai: Fudan University, 2008. | |
| [18] |
Jentsch S. The ubiquitin-conjugation system[J]. Annu Rev Genet, 1992, 26: 179-207.
pmid: 1336336 |
| [19] | Ahn MY, Oh TR, Seo DH, et al. Arabidopsis group XIV ubiquitin-conjugating enzymes AtUBC32, AtUBC33, and AtUBC34 play negative roles in drought stress response[J]. J Plant Physiol, 2018, 230: 73-79. |
| [20] | Jue DW, Sang XL, Liu LQ, et al. The ubiquitin-conjugating enzyme gene family in longan(Dimocarpus longan Lour.): genome-wide identification and gene expression during flower induction and abiotic stress responses[J]. Molecules, 2018, 23(3): 662. |
| [21] | Cui F, Liu LJ, Zhao QZ, et al. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance[J]. Plant Cell, 2012, 24(1): 233-244. |
| [22] | Wijk SJL, Marc Timmers HT. The family of ubiquitin-conjugating enzymes(E2s): deciding between life and death of proteins[J]. FASEB J, 2010, 24(4): 981-993. |
| [23] | 刘维刚. 马铃薯泛素结合酶E2基因家族鉴定和StUBC9基因克隆及功能研究[D]. 兰州: 甘肃农业大学, 2019. |
| Liu WG. Genome-wide identification of ubiquitin conjugating enzymes E2 gene family and cloning and functional aanlysis of StUBC9 in potato[D]. Lanzhou: Gansu Agricultural University, 2019. | |
| [24] | Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana[J]. Nature, 2000, 408(6814): 796-815. |
| [25] |
Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics[J]. Annu Rev Genomics Hum Genet, 2000, 1: 41-73.
pmid: 11701624 |
| [26] | 罗传英. 草莓泛素结合酶基因家族鉴定和调控果实成熟的功能初探[D]. 雅安: 四川农业大学, 2022. |
| Luo CY. Identification of ubiquitin conjugating enzyme gene family and functional analysis of regulating fruit ripening in strawberry[D]. Ya'an: Sichuan Agricultural University, 2022. | |
| [27] | Wang YY, Wang WH, Cai JH, et al. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening[J]. Genome Biol, 2014, 15(12): 548. |
| [28] | 王园, 王甲水, 谢学立, 等. 香蕉泛素结合酶基因与果实成熟关系的研究[J]. 园艺学报, 2010, 37(5): 705-712. |
| Wang Y, Wang JS, Xie XL, et al. Studies of the relationship between MaUCE1 and banana fruit ripening[J]. Acta Hortic Sin, 2010, 37(5): 705-712. | |
| [29] | Chung E, Cho CW, So HA, et al. Overexpression of VrUBC1, a mung bean E2 ubiquitin-conjugating enzyme, enhances osmotic stress tolerance in Arabidopsis[J]. PLoS One, 2013, 8(6): e66056. |
| [30] | Zhou GA, Chang RZ, Qiu LJ. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis[J]. Plant Mol Biol, 2010, 72(4/5): 357-367. |
| [31] | Wan XR, Mo AQ, Liu S, et al. Constitutive expression of a peanut ubiquitin-conjugating enzyme gene in Arabidopsis confers improved water-stress tolerance through regulation of stress-responsive gene expression[J]. J Biosci Bioeng, 2011, 111(4): 478-484. |
| [32] | Gong M, Wang H, Chen MJ, et al. A newly discovered ubiquitin-conjugating enzyme E2 correlated with the cryogenic autolysis of Volvariella volvacea[J]. Gene, 2016, 583(1): 58-63. |
| [1] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [2] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [3] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [4] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [5] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [6] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [7] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [8] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [9] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [10] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [11] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [12] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [13] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [14] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [15] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||