








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 172-182.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0094
Previous Articles Next Articles
SUN Hui-qiong( ), ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong(
), ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong( )
)
Received:2024-01-24
Online:2024-07-26
Published:2024-07-30
Contact:
JIANG Xiao-dong
E-mail:sunhuiqiong@163.com;sxaujiangxd@163.com
SUN Hui-qiong, ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong. Identification, Expression and DNA Variation Analysis of FLS Gene Family in Chenopodium quinoa[J]. Biotechnology Bulletin, 2024, 40(7): 172-182.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| CqFLS1.1g-qF | GGTCTTTGGTACAATGTCCGC |
| CqFLS1.1g-qR | GGGCCATGACATCCTTGTTTTT |
| EF1a-F | GACAAGCGTGTGATCGAGAG |
| EF1α-R | TCGGCCTTAAGTTTGTCGAGA |
| F1 | ATGGAGGTAGAAAAAGTGCAA |
| R1 | TTAACAAGCTCCCTCAGTGA |
Table 1 List of primer sequences
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| CqFLS1.1g-qF | GGTCTTTGGTACAATGTCCGC |
| CqFLS1.1g-qR | GGGCCATGACATCCTTGTTTTT |
| EF1a-F | GACAAGCGTGTGATCGAGAG |
| EF1α-R | TCGGCCTTAAGTTTGTCGAGA |
| F1 | ATGGAGGTAGAAAAAGTGCAA |
| R1 | TTAACAAGCTCCCTCAGTGA |
| 基因编号Gene code | 基因ID Gene ID | 染色体Chromosome | 染色体位置Genome location | 长度Length/ bp |
|---|---|---|---|---|
| CqFLS1.1g | AUR62004717 | Chr01 | 122 518 204-122 519 817 | 1 613 |
| CqFLS2.1g | AUR62014672 | Chr01 | 28 868 426-28 870 600 | 2 174 |
| CqFLS3.10g | AUR62023632 | Chr10 | 243 450-244 961 | 1 511 |
Table 2 Chenopodium quinoa chromosome location and gene structure
| 基因编号Gene code | 基因ID Gene ID | 染色体Chromosome | 染色体位置Genome location | 长度Length/ bp |
|---|---|---|---|---|
| CqFLS1.1g | AUR62004717 | Chr01 | 122 518 204-122 519 817 | 1 613 |
| CqFLS2.1g | AUR62014672 | Chr01 | 28 868 426-28 870 600 | 2 174 |
| CqFLS3.10g | AUR62023632 | Chr10 | 243 450-244 961 | 1 511 |
| 蛋白名称 Protein name | 氨基酸数量 Number of amino acids | 分子量 Molecular weight | 等电点 Theoretical pI | 不稳定指数Instability index | 脂肪族指数 Aliphatic index | 亲疏水性(GRAVY) Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| CqFLS3.10g | 346 | 39496.05 | 6.04 | 44.05 | 85.61 | -0.544 |
| CqFLS2.1g | 339 | 38423.03 | 6.28 | 42.27 | 85.40 | -0.492 |
| CqFLS1.1g | 346 | 39409.91 | 6.04 | 46.50 | 84.48 | -0.549 |
Table 3 Physical and chemical characteristics of CqFLSs
| 蛋白名称 Protein name | 氨基酸数量 Number of amino acids | 分子量 Molecular weight | 等电点 Theoretical pI | 不稳定指数Instability index | 脂肪族指数 Aliphatic index | 亲疏水性(GRAVY) Grand average of hydropathicity |
|---|---|---|---|---|---|---|
| CqFLS3.10g | 346 | 39496.05 | 6.04 | 44.05 | 85.61 | -0.544 |
| CqFLS2.1g | 339 | 38423.03 | 6.28 | 42.27 | 85.40 | -0.492 |
| CqFLS1.1g | 346 | 39409.91 | 6.04 | 46.50 | 84.48 | -0.549 |
| 蛋白名称 Protein name | α-螺旋 Alpha helix/% | 延伸链 Extended strand/% | β-折叠 Beta turn/% | 无规则卷曲 Random coil/% | 亚细胞定位 Intracellular location/% | 可能性 Probabily/% |
|---|---|---|---|---|---|---|
| CqFLS3.10g | 31.79 | 18.21 | 5.78 | 44.22 | Cytoplasmic | 65.2 |
| CqFLS2.1g | 33.92 | 17.99 | 5.60 | 42.48 | Cytoplasmic | 65.2 |
| CqFLS1.1g | 32.95 | 19.65 | 6.36 | 41.04 | Cytoplasmic | 65.2 |
Table 4 Secondary structure and subcellular localization of CqFLSs
| 蛋白名称 Protein name | α-螺旋 Alpha helix/% | 延伸链 Extended strand/% | β-折叠 Beta turn/% | 无规则卷曲 Random coil/% | 亚细胞定位 Intracellular location/% | 可能性 Probabily/% |
|---|---|---|---|---|---|---|
| CqFLS3.10g | 31.79 | 18.21 | 5.78 | 44.22 | Cytoplasmic | 65.2 |
| CqFLS2.1g | 33.92 | 17.99 | 5.60 | 42.48 | Cytoplasmic | 65.2 |
| CqFLS1.1g | 32.95 | 19.65 | 6.36 | 41.04 | Cytoplasmic | 65.2 |

Fig. 3 Analysis of CqFLS proteins A: Domain analysis of CqFLS proteins; B: conserved motif analysis of CqFLS proteins; C: protein sequence alignment analysis of CqFLS
| 基因编号Gene code | 位置POS | 参考REF | 变异ALT | 品系Q3 | 品系Q6 | 品系Q8 | 品系Q9 | 影响Effect |
|---|---|---|---|---|---|---|---|---|
| CqFLS2.1g | 28871893 | CT | C | C | N | C | C | UPSTREAM |
| CqFLS2.1g | 28871125 | TA | T | N | T | T | T | UPSTREAM |
| CqFLS2.1g | 28872881 | TTA | T | N | N | N | T | UPSTREAM |
| CqFLS2.1g | 28872134 | T | A | A | A | A | N | UPSTREAM |
| CqFLS2.1g | 28871765 | C | T | T | N | T | T | UPSTREAM |
| CqFLS2.1g | 28871453 | T | G | G | G | G | G | UPSTREAM |
| CqFLS2.1g | 28870264 | C | T | T | T | T | N | SYNONYMOUS_CODING |
| CqFLS2.1g | 28871914 | A | T | T | N | T | T | UPSTREAM |
| CqFLS2.1g | 28871369 | G | A | A | A | A | A | UPSTREAM |
| CqFLS2.1g | 28870706 | T | G | G | N | G | N | UPSTREAM |
| CqFLS2.1g | 28872268 | T | C | C | C | N | C | UPSTREAM |
| CqFLS2.1g | 28872086 | G | A | A | A | A | N | UPSTREAM |
| CqFLS2.1g | 28871206 | G | T | T | N | T | T | UPSTREAM |
Table 5 DNA variation of CqFLSs among quinoa lines
| 基因编号Gene code | 位置POS | 参考REF | 变异ALT | 品系Q3 | 品系Q6 | 品系Q8 | 品系Q9 | 影响Effect |
|---|---|---|---|---|---|---|---|---|
| CqFLS2.1g | 28871893 | CT | C | C | N | C | C | UPSTREAM |
| CqFLS2.1g | 28871125 | TA | T | N | T | T | T | UPSTREAM |
| CqFLS2.1g | 28872881 | TTA | T | N | N | N | T | UPSTREAM |
| CqFLS2.1g | 28872134 | T | A | A | A | A | N | UPSTREAM |
| CqFLS2.1g | 28871765 | C | T | T | N | T | T | UPSTREAM |
| CqFLS2.1g | 28871453 | T | G | G | G | G | G | UPSTREAM |
| CqFLS2.1g | 28870264 | C | T | T | T | T | N | SYNONYMOUS_CODING |
| CqFLS2.1g | 28871914 | A | T | T | N | T | T | UPSTREAM |
| CqFLS2.1g | 28871369 | G | A | A | A | A | A | UPSTREAM |
| CqFLS2.1g | 28870706 | T | G | G | N | G | N | UPSTREAM |
| CqFLS2.1g | 28872268 | T | C | C | C | N | C | UPSTREAM |
| CqFLS2.1g | 28872086 | G | A | A | A | A | N | UPSTREAM |
| CqFLS2.1g | 28871206 | G | T | T | N | T | T | UPSTREAM |
| 基因编号 Gene code | 非同义突变率 Ka | 非同义突变率 Ks | 比值 Ka/Ks |
|---|---|---|---|
| CqFLS3.10g | 0.104881 | 0.726108 | 0.144443 |
| CqFLS2.1g | 0.140285 | 0.742346 | 0.188976 |
| CqFLS1.1g | 0.102561 | 0.767349 | 0.133656 |
Table 6 Evolutionary selection parameters for members of the CqFLSs
| 基因编号 Gene code | 非同义突变率 Ka | 非同义突变率 Ks | 比值 Ka/Ks |
|---|---|---|---|
| CqFLS3.10g | 0.104881 | 0.726108 | 0.144443 |
| CqFLS2.1g | 0.140285 | 0.742346 | 0.188976 |
| CqFLS1.1g | 0.102561 | 0.767349 | 0.133656 |
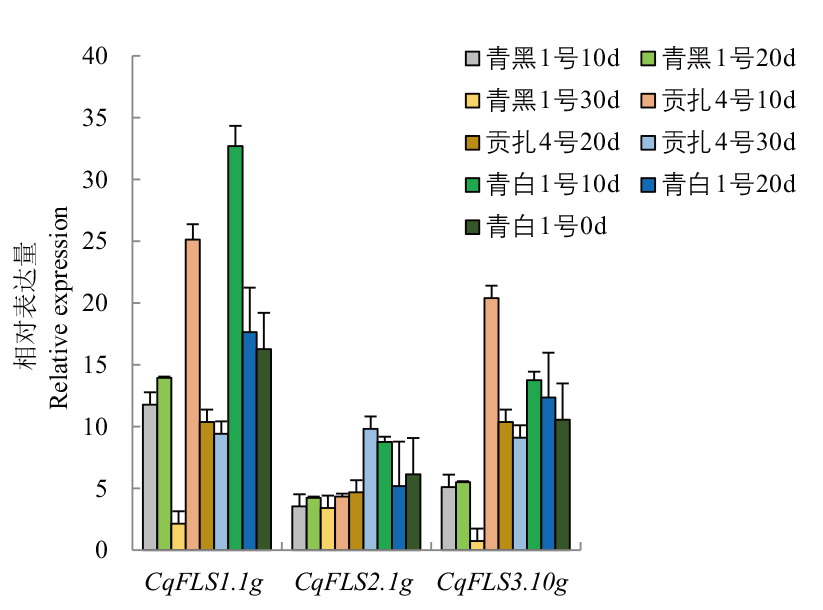
Fig. 7 Expressions of FLS in different varieties of quinoa seeds 10 d, 20 d, 30 d refer to 10 days 20 days and 30 days after pollination, respectively

Fig. 9 SDS-PAGE analysis of CqFLS1.1g prokaryotic expression(A)and protein purification(B) (A)M: Protein marker; 1: pET-28a(+)pre-induction total protein; 2: pET-28a(+)vector control; 3: PET-28a-CqFLS1.1g was induced without IPTG; 4: IPTG-induced supernatant at 20℃; 5: IPTG-induced precipitation at 20℃; 6: IPTG-induced supernatant at 37℃; 7: IPTG induces precipitation at 37℃.(B)M: Protein marker; 1: final purified target protein
| [1] | 丰扬, 郭凤根, 王仕玉, 等. 藜麦Cq6GT基因的克隆与表达分析[J]. 植物生理学报, 2022, 58(10): 2017-2024. |
| Feng Y, Guo FG, Wang SY, et al. Cloning and expression analysis of Cq6GT gene from Chenopodium quinoa[J]. Plant Physiol J, 2022, 58(10): 2017-2024. | |
| [2] | Nowak V, Du J, Charrondière UR. Assessment of the nutritional composition of quinoa(Chenopodium quinoa Willd.)[J]. Food Chem, 2016, 193: 47-54. |
| [3] | Li XH, Kim YB, Kim Y, et al. Differential stress-response expression of two flavonol synthase genes and accumulation of flavonols in Tartary buckwheat[J]. J Plant Physiol, 2013, 170(18): 1630-1636. |
| [4] | Kou M, Li C, Song WH, et al. Identification and functional characterization of a flavonol synthase gene from sweet potato[Ipomoea batatas(L.) Lam.][J]. Front Plant Sci, 2023, 14: 1181173. |
| [5] | Kimura S, Nakatsuka T, Yamada E, et al. A flavonol synthase gene GtFLS defines anther-specific flavonol accumulation in gentian[J]. Plant Biotechnol, 2010, 28(2): 211-221. |
| [6] |
Harborne JB, Williams CA. Advances in flavonoid research since 1992[J]. Phytochemistry, 2000, 55(6): 481-504.
doi: 10.1016/s0031-9422(00)00235-1 pmid: 11130659 |
| [7] | Deis L, Cavagnaro B, Bottini R, et al. Water deficit and exogenous ABA significantly affect grape and wine phenolic composition under in field and in-vitro conditions[J]. Plant Growth Regul, 2011, 65(1): 11-21. |
| [8] | Iwashina T. The structure and distribution of the flavonoids in plants[J]. J Plant Res, 2000, 113(3): 287-299. |
| [9] | Ishikura N, Yoshitama K. Anthocyanin-flavonol co-pigmentation in blue seed Coats of Ophiopogon jaburan[J]. J Plant Physiol, 1984, 115(2): 171-175. |
| [10] | Hichri F, Ben Salah N, Omri A, et al. New antioxidant C-glycosyl flavone and flavonol derivatives from the Tunisian Achille acretica L[J]. S Afr N J Bot, 2018, 116: 1-5. |
| [11] | Marzouk MS, Moharram FA, Haggag EG, et al. Antioxidant flavonol glycosides from Schinus molle[J]. Phytother Res, 2006, 20(3): 200-205. |
| [12] | Corsino J, Silva DHS, Zanoni MVB, et al. Antioxidant flavan-3-ols and flavonol glycosides from Maytenus aquifolium[J]. Phytother Res, 2003, 17(8): 913-916. |
| [13] |
Dias TA, Duarte CL, Lima CF, et al. Superior anticancer activity of halogenated chalcones and flavonols over the natural flavonol quercetin[J]. Eur J Med Chem, 2013, 65: 500-510.
doi: 10.1016/j.ejmech.2013.04.064 pmid: 23771043 |
| [14] |
Britton RG, Horner-Glister E, Pomenya OA, et al. Synthesis and biological evaluation of novel flavonols as potential anti-prostate cancer agents[J]. Eur J Med Chem, 2012, 54: 952-958.
doi: 10.1016/j.ejmech.2012.06.031 pmid: 22789812 |
| [15] |
Li SG, Dong P, Wang JW, et al. Icariin, a natural flavonol glycoside, induces apoptosis in human hepatoma SMMC-7721 cells via a ROS/JNK-dependent mitochondrial pathway[J]. Cancer Lett, 2010, 298(2): 222-230.
doi: 10.1016/j.canlet.2010.07.009 pmid: 20674153 |
| [16] | Ortega YH, Foubert K, Vanden Berghe W, et al. Flavonol glycosides from the leaves of Boldoa purpurascens and their anti-inflammatory properties[J]. Phytochem Lett, 2017, 19: 71-76. |
| [17] | Tahiri O, Atmani-Kilani D, Sanchez-Fidalgo S, et al. The flavonol-enriched Cistus albidus chloroform extract possesses in vivo anti-inflammatory and anti-nociceptive activity[J]. J Ethnopharmacol, 2017, 209: 210-218. |
| [18] | Granica S, Czerwińska ME, Żyżyńska-Granica B, et al. Antioxidant and anti-inflammatory flavonol glucuronides from Polygonum aviculare L[J]. Fitoterapia, 2013, 91: 180-188. |
| [19] |
Şöhretoğlu D, Sari S, Barut B, et al. Discovery of potent α-glucosidase inhibitor flavonols: insights into mechanism of action through inhibition kinetics and docking simulations[J]. Bioorg Chem, 2018, 79: 257-264.
doi: S0045-2068(18)30332-8 pmid: 29778797 |
| [20] | Sendrayaperumal V, Iyyam Pillai S, Subramanian S. Design, synthesis and characterization of zinc-morin, a metal flavonol complex and evaluation of its antidiabetic potential in HFD-STZ induced type 2 diabetes in rats[J]. Chem Biol Interact, 2014, 219: 9-17. |
| [21] | Sun YJ, He JM, Kong JQ. Characterization of two flavonol synthases with iron-independent flavanone 3-hydroxylase activity from Ornithogalum caudatum Jacq[J]. BMC Plant Biol, 2019, 19(1): 195. |
| [22] |
Britsch L, Dedio J, Saedler H, et al. Molecular characterization of flavanone 3 beta-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues[J]. Eur J Biochem, 1993, 217(2): 745-754.
pmid: 8223617 |
| [23] | Kim YB, Kim K, Kim Y, et al. Cloning and characterization of a flavonol synthase gene from Scutellaria baicalensis[J]. Sci World J, 2014, 2014: 980740. |
| [24] | Kim BG, Joe EJ, Ahn JH. Molecular characterization of flavonol synthase from poplar and its application to the synthesis of 3-O-methylkaempferol[J]. Biotechnol Lett, 2010, 32(4): 579-584. |
| [25] | Falcone Ferreyra ML, Rius S, Emiliani J, et al. Cloning and characterization of a UV-B-inducible maize flavonol synthase[J]. Plant J, 2010, 62(1): 77-91. |
| [26] | Li CL, Bai YC, Li SJ, et al. Cloning, characterization, and activity analysis of a flavonol synthase gene FtFLS1 and its association with flavonoid content in Tartary buckwheat[J]. J Agric Food Chem, 2012, 60(20): 5161-5168. |
| [27] | Xu F, Li LL, Zhang WW, et al. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba[J]. Mol Biol Rep, 2012, 39(3): 2285-2296. |
| [28] | Artimo P, Jonnalagedda M, Arnold K, et al. ExPASy: SIB bioinformatics resource portal[J]. Nucleic Acids Res, 2012, 40(Web Server issue): W597-W603. |
| [29] | Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Comput Appl Biosci, 1995, 11(6): 681-684. |
| [30] |
Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks[J]. Nat Biotechnol, 2019, 37(4): 420-423.
doi: 10.1038/s41587-019-0036-z pmid: 30778233 |
| [31] | 姜晓东, 李新凤, 郝艳平, 等. 藜麦β-香树酯醇合酶和鲨烯合酶基因的克隆与表达[J]. 土壤, 2018, 50(6): 1214-1221. |
| Jiang XD, Li XF, Hao YP, et al. Gene cloning and express of squalene synthase and β-amyrin synthase from Chenopodium quinoa[J]. Soils, 2018, 50(6): 1214-1221. | |
| [32] |
Dooner HK, Robbins TP, Jorgensen RA. Genetic and developmental control of anthocyanin biosynthesis[J]. Annu Rev Genet, 1991, 25: 173-199.
pmid: 1839877 |
| [33] | Balakrishnan G, Schneider RG. Quinoa flavonoids and their bioaccessibility during in vitro gastrointestinal digestion[J]. J Cereal Sci, 2020, 95: 103070. |
| [34] |
Pelletier MK, Murrell JR, Shirley BW. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of “early” and “late” genes[J]. Plant Physiol, 1997, 113(4): 1437-1445.
pmid: 9112784 |
| [35] | Liu YJ, Liu JN, Kong ZY, et al. Transcriptomics and metabolomics analyses of the mechanism of flavonoid synthesis in seeds of differently colored quinoa strains[J]. Genomics, 2022, 114(1): 138-148. |
| [36] | 宋奇琦, 张小秋, 宋修鹏, 等. 甘蔗HSP20基因克隆、原核表达及逆境胁迫响应[J]. 植物生理学报, 2022, 58(2): 371-380. |
| Song QQ, Zhang XQ, Song XP, et al. Cloning and prokaryotic expression of sugarcane HSP20 gene and its responses to adversity stress[J]. Plant Physiol J, 2022, 58(2): 371-380. |
| [1] | ZHANG Na, LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong. Alternative Translation of Wheat Enolase-encoding Gene ENO2 and Its Prokaryotic Expression [J]. Biotechnology Bulletin, 2024, 40(5): 112-119. |
| [2] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [3] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [4] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [5] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [6] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [7] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [8] | WANG Guang-li, FAN Chan, WANG Hui, LU Hui-fang, XIA Ling-yin, HUANG Jian, MIN Xun. Prokaryotic Expression,Purification,Identification,and Polyclonal Antibody Preparation of Vibrio cholerae Hemolysin HlyA [J]. Biotechnology Bulletin, 2022, 38(7): 269-277. |
| [9] | SUN Bao-zhen, QUAN Long-ping, KANG Hui, YAO Yu-xin, SHEN Tian, CHEN Wei-ping, DU Yuan-peng, GAO Zhen. Back-splicing Primers-based PCR Method for Specific Detection of circRNA [J]. Biotechnology Bulletin, 2022, 38(5): 279-285. |
| [10] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [11] | SHEN Jun-qiang, ZHANG Li-ping, YU Rui-ming, WANG Yong-lu, PAN Li, LIU Xia, LIU Xin-sheng. Porcine Kobuvirus Structural Proteins VP0 and VP1 Prokaryotic Expression and Establishment of Indirect ELISA Method [J]. Biotechnology Bulletin, 2022, 38(10): 243-253. |
| [12] | FAN Ya-peng, RUI Cun, ZHANG Yue-xin, CHEN Xiu-gui, LU Xu-ke, WANG Shuai, ZHANG Hong, XU Nan, WANG Jing, CHEN Chao, YE Wu-wei. Cloning,Expression and Preliminary Bioinformatics Analysis of the Alkaline Tolerant Gene GhZAT12 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 121-130. |
| [13] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [14] | ZENG Fu-yuan, SU Ze-hui, ZHOU Shi-hui, XIE Miao, PANG Huan-ying. Prokaryotic Expression of the PEPCK Protein of Vibrio alginolyticus and Identification of Its Acetylation and Succinylation [J]. Biotechnology Bulletin, 2021, 37(5): 84-91. |
| [15] | ZHANG Xi-xi, ZHANG Yi-qing, LI Yu-lin, HAN Xiao, WANG Guo-qiang, WANG Xiao-jun, WANG Xu-dong, WANG Yun-long. Prokaryotic Expression,Purification and Application of N Protein C-terminal Recombinant Protein in Novel Coronavirus(SARS-CoV-2) [J]. Biotechnology Bulletin, 2021, 37(5): 92-97. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||