








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 163-171.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0162
Previous Articles Next Articles
ZANG Wen-rui( ), MA Ming, CHE Gen, HASI Agula(
), MA Ming, CHE Gen, HASI Agula( )
)
Received:2024-02-18
Online:2024-07-26
Published:2024-07-30
Contact:
HASI Agula
E-mail:1211589256@qq.com;hasiagula@imu.edu.cn
ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon[J]. Biotechnology Bulletin, 2024, 40(7): 163-171.
| 基因名称 Gene name | 登录号Accession No. | 染色体分布Chromosome distribution | 染色体座位Chromosomal loci/bp | ORF /bp | 氨基酸数量Number of amino acids | 分子量Molecular weight/kD | 等电点Isoelectric point(pI) |
|---|---|---|---|---|---|---|---|
| CmBZR1 | MELO3C010925.2.1 | Chr 03 | 29999092-30000475 | 936 | 311 | 34.05 | 9.16 |
| CmBES1 | MELO3C007804.2.1 | Chr 08 | 5468519-5471484 | 960 | 319 | 34.71 | 8.96 |
| CmBEH1 | MELO3C016213.2.1 | Chr 07 | 22268773-22275513 | 1 932 | 643 | 75.11 | 5.90 |
| CmBEH2 | MELO3C021214.2.1 | Chr 11 | 31280641-31285843 | 2 046 | 681 | 78.21 | 5.74 |
| CmBEH3 | MELO3C016121.2.1 | Chr 07 | 20822745-20826732 | 978 | 325 | 34.70 | 8.50 |
| CmBEH4 | MELO3C002681.2.1 | Chr 12 | 22058946-22061841 | 984 | 327 | 39.52 | 9.00 |
Table 1 Basic information of BZR family members in melon
| 基因名称 Gene name | 登录号Accession No. | 染色体分布Chromosome distribution | 染色体座位Chromosomal loci/bp | ORF /bp | 氨基酸数量Number of amino acids | 分子量Molecular weight/kD | 等电点Isoelectric point(pI) |
|---|---|---|---|---|---|---|---|
| CmBZR1 | MELO3C010925.2.1 | Chr 03 | 29999092-30000475 | 936 | 311 | 34.05 | 9.16 |
| CmBES1 | MELO3C007804.2.1 | Chr 08 | 5468519-5471484 | 960 | 319 | 34.71 | 8.96 |
| CmBEH1 | MELO3C016213.2.1 | Chr 07 | 22268773-22275513 | 1 932 | 643 | 75.11 | 5.90 |
| CmBEH2 | MELO3C021214.2.1 | Chr 11 | 31280641-31285843 | 2 046 | 681 | 78.21 | 5.74 |
| CmBEH3 | MELO3C016121.2.1 | Chr 07 | 20822745-20826732 | 978 | 325 | 34.70 | 8.50 |
| CmBEH4 | MELO3C002681.2.1 | Chr 12 | 22058946-22061841 | 984 | 327 | 39.52 | 9.00 |

Fig. 1 Analysis of CmBZR protein sequences A: Multiple sequence alignment of 6 CmBZR proteins; B: conserved sites of BZR domains in CmBZR sequences. *: Conserved motif of CmBZR protein sequences
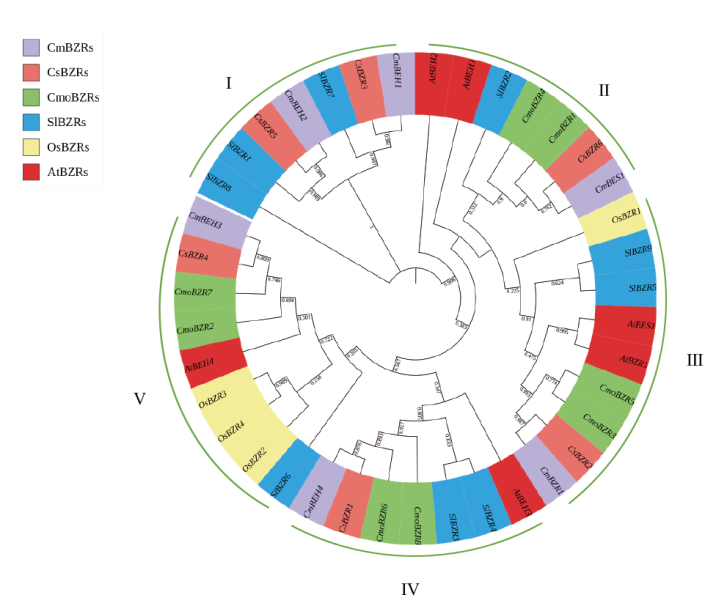
Fig. 3 Phylogenetic tree analysis of BZR proteins from Arabidopsis,Oryza sativa,Solanum lycopersicon,Cucumis sativus,Cucurbita moschata and C. smelo I-V indicates different subfamily of BZR members in six species
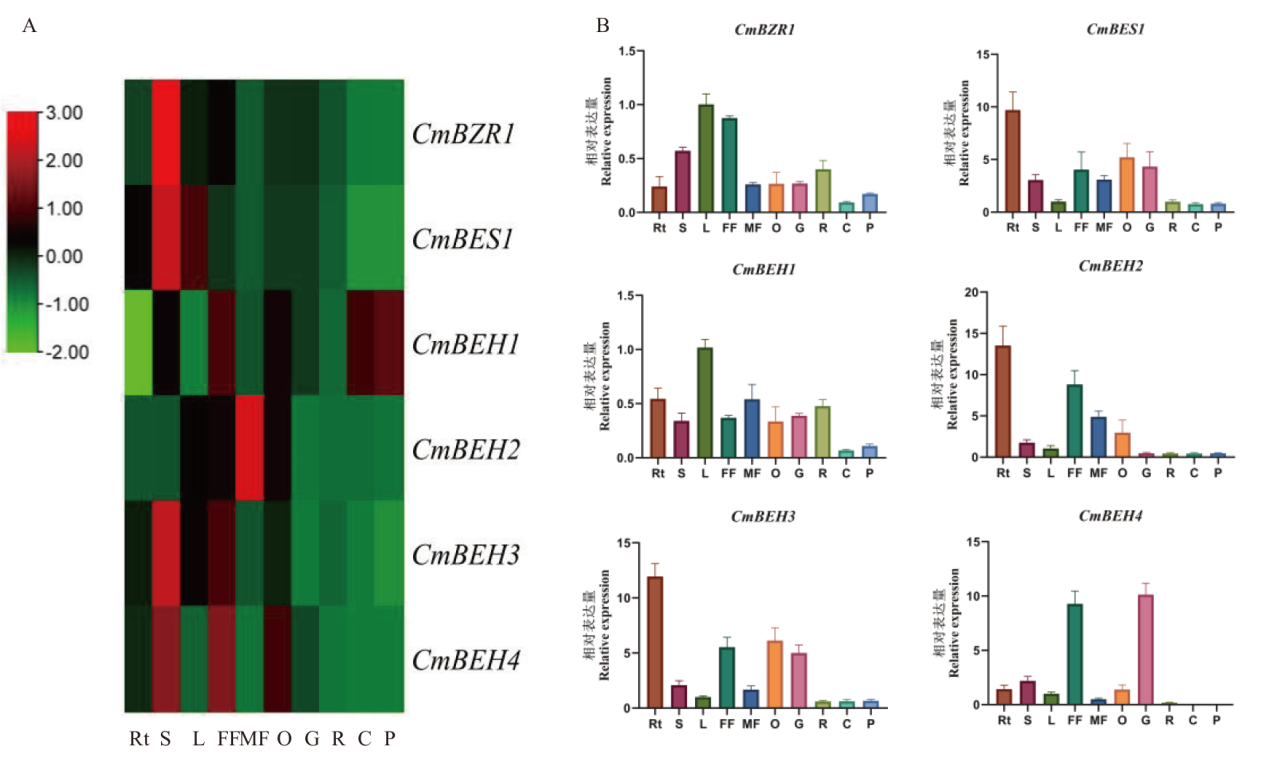
Fig. 5 Expression analysis of CmBZR in melon A: Heat map of 6 CmBZR genes expressed differently in different tissues of melon;B: expression profiles of CmBZR genes in melon’s Rt, S, L, FF, MF, O, G,R, C and P, and they indicate the root, stem, leaf, female flower, male flower, ovary, growing stage, ripening stage, climacteric stage, and post-climacteric stage fruit, respectively.
| [1] | Shin AY, Kim YM, Koo N, et al. Transcriptome analysis of the oriental melon(Cucumis melo L. var. makuwa)during fruit development[J]. PeerJ, 2017, 5: e2834. |
| [2] | Yin YH, Vafeados D, Tao Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis[J]. Cell, 2005, 120(2): 249-259. |
| [3] |
Steber CM, McCourt P. A role for brassinosteroids in germination in Arabidopsis[J]. Plant Physiol, 2001, 125(2): 763-769.
doi: 10.1104/pp.125.2.763 pmid: 11161033 |
| [4] | Yang CJ, Zhang C, Lu YN, et al. The mechanisms of brassinosteroids'action: from signal transduction to plant development[J]. Mol Plant, 2011, 4(4): 588-600. |
| [5] | Galstyan A, Nemhauser JL. Auxin promotion of seedling growth via ARF5 is dependent on the brassinosteroid-regulated transcription factors BES1 and BEH4[J]. Plant Direct, 2019, 3(9): e00166. |
| [6] |
Liang T, Mei SL, Shi C, et al. UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis[J]. Dev Cell, 2018, 44(4): 512-523.e5.
doi: S1534-5807(17)31079-1 pmid: 29398622 |
| [7] | Wang WX, Lu XD, Li L, et al. Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis[J]. Plant Cell, 2018, 30(9): 1989-2005. |
| [8] | Bajguz A, Hayat S. Effects of brassinosteroids on the plant responses to environmental stresses[J]. Plant Physiol Biochem, 2009, 47(1): 1-8. |
| [9] |
Liao K, Peng YJ, Yuan LB, et al. Brassinosteroids antagonize jasmonate-activated plant defense responses through BRI1-EMS-SUPPRESSOR1(BES1)[J]. Plant Physiol, 2020, 182(2): 1066-1082.
doi: 10.1104/pp.19.01220 pmid: 31776183 |
| [10] | Cui XY, Gao Y, Guo J, et al. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1[J]. Plant Physiol, 2019, 180(1): 605-620. |
| [11] | Jaillais Y, Hothorn M, Belkhadir Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor[J]. Genes Dev, 2011, 25(3): 232-237. |
| [12] | Tang WQ, Kim TW, Oses-Prieto JA, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis[J]. Science, 2008, 321(5888): 557-560. |
| [13] | Kim TW, Guan SH, Burlingame AL, et al. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2[J]. Mol Cell, 2011, 43(4): 561-571. |
| [14] |
Peng P, Yan ZY, Zhu YY, et al. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation[J]. Mol Plant, 2008, 1(2): 338-346.
doi: 10.1093/mp/ssn001 pmid: 18726001 |
| [15] | Tang WQ, Yuan M, Wang RJ, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1[J]. Nat Cell Biol, 2011, 13(2): 124-131. |
| [16] |
Yin YH, Wang ZY, Mora-Garcia S, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation[J]. Cell, 2002, 109(2): 181-191.
doi: 10.1016/s0092-8674(02)00721-3 pmid: 12007405 |
| [17] |
Wang ZY, Nakano T, Gendron J, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis[J]. Dev Cell, 2002, 2(4): 505-513.
doi: 10.1016/s1534-5807(02)00153-3 pmid: 11970900 |
| [18] | Bai MY, Zhang LY, Gampala SS, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice[J]. Proc Natl Acad Sci USA, 2007, 104(34): 13839-13844. |
| [19] | Gampala SS, Kim TW, He JX, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis[J]. Dev Cell, 2007, 13(2): 177-189. |
| [20] |
Szekeres M, Németh K, Koncz-Kálmán Z, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis[J]. Cell, 1996, 85(2): 171-182.
doi: 10.1016/s0092-8674(00)81094-6 pmid: 8612270 |
| [21] |
Gil P, Liu Y, Orbović V, et al. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis[J]. Plant Physiol, 1994, 104(2): 777-784.
doi: 10.1104/pp.104.2.777 pmid: 8159792 |
| [22] | Xie LQ, Yang CJ, Wang XL. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis[J]. J Exp Bot, 2011, 62(13): 4495-4506. |
| [23] |
Ibañez C, Delker C, Martinez C, et al. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1[J]. Curr Biol, 2018, 28(2): 303-310.e3.
doi: S0960-9822(17)31602-0 pmid: 29337075 |
| [24] |
Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses[J]. Nat Cell Biol, 2012, 14(8): 802-809.
doi: 10.1038/ncb2545 pmid: 22820378 |
| [25] | Chen LG, Gao ZH, Zhao ZY, et al. BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis[J]. Mol Plant, 2019, 12(10): 1408-1415. |
| [26] | 于好强, 孙福艾, 冯文奇, 等. 转录因子BES1/BZR1调控植物生长发育及抗逆性[J]. 遗传, 2019, 41(3): 206-214. |
| Yu HQ, Sun FA, Feng WQ, et al. The BES1/BZR1 transcription factors regulate growth, development and stress resistance in plants[J]. Hereditas, 2019, 41(3): 206-214. | |
| [27] | He JX, Gendron JM, Sun Y, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses[J]. Science, 2005, 307(5715): 1634-1638. |
| [28] | Luo SL, Zhang GB, Zhang ZY, et al. Genome-wide identification and expression analysis of BZR gene family and associated responses to abiotic stresses in cucumber(Cucumis sativus L.)[J]. BMC Plant Biol, 2023, 23(1): 214. |
| [29] | 李春, 刘小俊, 蔡鹏, 等. 中国南瓜BZR基因家族的全基因组鉴定及生物信息学分析[J]. 分子植物育种, 2022, 20(19): 6324-6330. |
| Li C, Liu XJ, Cai P, et al. Genome-wide identification and bioinformatics analysis of BZR gene family in pumpkin(Cucurbita moschata duch.)[J]. Mol Plant Breed, 2022, 20(19): 6324-6330. | |
| [30] | 陈旭, 沈春洋, 莫福磊, 等. 番茄BZR基因家族鉴定及非生物胁迫下表达模式分析[J]. 东北农业大学学报, 2021, 52(11): 9-17. |
| Chen X, Shen CY, Mo FL, et al. Identification of BZR gene family in tomato and expression patterns analysis under abiotic stress[J]. J Northeast Agric Univ, 2021, 52(11): 9-17. | |
| [31] | Zheng Y, Wu S, Bai Y, et al. Cucurbit Genomics Database(CuGenDB): a central portal for comparative and functional genomics of cucurbit crops[J]. Nucleic Acids Res, 2019, 47(D1): D1128-D1136. |
| [32] |
Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure[J]. BMC Bioinformatics, 2010, 11: 431.
doi: 10.1186/1471-2105-11-431 pmid: 20718988 |
| [33] | Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020[J]. Nucleic Acids Res, 2021, 49(D1): D458-D460. |
| [34] |
Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server[J]. Methods Mol Biol, 1999, 112: 531-552.
pmid: 10027275 |
| [35] |
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11[J]. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [36] |
Waterhouse AM, Procter JB, Martin DMA, et al. Jalview Version 2—a multiple sequence alignment editor and analysis workbench[J]. Bioinformatics, 2009, 25(9): 1189-1191.
doi: 10.1093/bioinformatics/btp033 pmid: 19151095 |
| [37] |
Crooks GE, Hon G, Chandonia JM, et al. WebLogo: a sequence logo generator[J]. Genome Res, 2004, 14(6): 1188-1190.
doi: 10.1101/gr.849004 pmid: 15173120 |
| [38] |
Bolser D, Staines DM, Pritchard E, et al. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data[J]. Methods Mol Biol, 2016, 1374: 115-140.
doi: 10.1007/978-1-4939-3167-5_6 pmid: 26519403 |
| [39] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [40] | 江倩倩, 王雨婷, 惠竹梅. 葡萄BZR基因家族的鉴定及表达分析[J]. 植物生理学报, 2021, 57(6): 1218-1228. |
| Jiang QQ, Wang YT, Hui ZM. Identification and expression analysis of BZR gene family in grapevine[J]. Plant Physiol J, 2021, 57(6): 1218-1228. | |
| [41] | Manoli A, Trevisan S, Quaggiotti S, et al. Identification and characterization of the BZR transcription factor family and its expression in response to abiotic stresses in Zea mays L[J]. Plant Growth Regul, 2018, 84(3): 423-436. |
| [42] | Sarwar R, Geng R, Li L, et al. Genome-wide prediction, functional divergence, and characterization of stress-responsive BZR transcription factors in B. napus[J]. Front Plant Sci, 2022, 12: 790655. |
| [43] | Li YY, He LL, Li J, et al. Genome-wide identification, characterization, and expression profiling of the legume BZR transcription factor gene family[J]. Front Plant Sci, 2018, 9: 1332. |
| [44] | Cao X, Khaliq A, Lu S, et al. Genome-wide identification and characterization of the BES1 gene family in apple(Malus domestica)[J]. Plant Biol, 2020, 22(4): 723-733. |
| [45] | Song XM, Ma X, Li CJ, et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species[J]. BMC Genomics, 2018, 19(1): 346. |
| [1] | ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum [J]. Biotechnology Bulletin, 2024, 40(7): 150-162. |
| [2] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [3] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [4] | WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors [J]. Biotechnology Bulletin, 2024, 40(6): 23-33. |
| [5] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [6] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [7] | ZHANG Di, JU Rui, LI Li-mei, WANG Yu-qian, CHEN Rui, WANG Xin-yi. Application of Transcription Factor-based Biosensors in Environmental Analysis [J]. Biotechnology Bulletin, 2024, 40(6): 114-125. |
| [8] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [9] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [10] | WANG Qiu-yue, DUAN Peng-liang, LI Hai-xiao, LIU Ning, CAO Zhi-yan, DONG Jin-gao. Construction of cDNA Library of Setosphaeria turcica and Screening of Transcription Factor StMR1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(6): 281-289. |
| [11] | ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia [J]. Biotechnology Bulletin, 2024, 40(6): 180-189. |
| [12] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [13] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [14] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [15] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||