








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 313-320.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0454
Previous Articles Next Articles
MENG Ya-qi1( ), WANG Song2, YANG Peng1, YU Hang1, YAO Xu-dong1, GUO Yan-hua1, TANG Hong1, ZHANG Yi-yuan1, WANG Li-min1(
), WANG Song2, YANG Peng1, YU Hang1, YAO Xu-dong1, GUO Yan-hua1, TANG Hong1, ZHANG Yi-yuan1, WANG Li-min1( ), ZHOU Ping1(
), ZHOU Ping1( )
)
Received:2025-05-03
Online:2025-10-26
Published:2025-10-28
Contact:
WANG Li-min, ZHOU Ping
E-mail:1505506744@qq.com;wanglm1980@126.com;zhpxqf@163.com
MENG Ya-qi, WANG Song, YANG Peng, YU Hang, YAO Xu-dong, GUO Yan-hua, TANG Hong, ZHANG Yi-yuan, WANG Li-min, ZHOU Ping. Precise Editing of the FGF18 Gene in Sheep Fibroblasts Using the AncBE4max System[J]. Biotechnology Bulletin, 2025, 41(10): 313-320.
名称 Name | 外显子区域 Exon region | 引物序列 Primer sequence (5'-3') |
|---|---|---|
| sg1 | 第三外显子 | F:accgGGAGAACCAGACGCGGGCTC |
| R:aaacGAGCCCGCGTCTGGTTCTCC | ||
| sg2 | 第三外显子 | F:accgACATCCAGGTCCTGGGCCGC |
| R:aaacGCGGCCCAGGACCTGGATGT | ||
| sg3 | 第四外显子 | F:accgGGTAGTCAAGTCCGGATCAA |
| R:aaacTTGATCCGGACTTGACTACC |
Table 1 sgRNAs’ sequence
名称 Name | 外显子区域 Exon region | 引物序列 Primer sequence (5'-3') |
|---|---|---|
| sg1 | 第三外显子 | F:accgGGAGAACCAGACGCGGGCTC |
| R:aaacGAGCCCGCGTCTGGTTCTCC | ||
| sg2 | 第三外显子 | F:accgACATCCAGGTCCTGGGCCGC |
| R:aaacGCGGCCCAGGACCTGGATGT | ||
| sg3 | 第四外显子 | F:accgGGTAGTCAAGTCCGGATCAA |
| R:aaacTTGATCCGGACTTGACTACC |
名称 Name | 引物序列 Primer sequence (5'-3') | 退火温度 Annealing temperature (℃) | 产物长度 Product size (bp) |
|---|---|---|---|
| E3-1 | F:GCCAAGCAGGGCAGTTAC | 57.9 | 562 |
| R:GGCATTGACCAGCAAGAGTA | |||
| E4-1 | F:GAGTTGGGAACAGGTGTCA | 55.6 | 588 |
| R:CTGCTAAGCAAGCCAGAGT |
Table 2 PCR primers' sequence
名称 Name | 引物序列 Primer sequence (5'-3') | 退火温度 Annealing temperature (℃) | 产物长度 Product size (bp) |
|---|---|---|---|
| E3-1 | F:GCCAAGCAGGGCAGTTAC | 57.9 | 562 |
| R:GGCATTGACCAGCAAGAGTA | |||
| E4-1 | F:GAGTTGGGAACAGGTGTCA | 55.6 | 588 |
| R:CTGCTAAGCAAGCCAGAGT |
名称 Name | 脱靶序列 Off-target sequence | PCR引物序列 PCR primers’ sequence (5'-3') | 染色体位置 Chromosomal location |
|---|---|---|---|
| 脱靶-1 | GGACAGACAGAGGCGGGCTC | F:TGATGGATGTTGGCAGTGGT R:GCTGACAGACGCCGTGGTTC | 18 |
| 脱靶-2 | CCAGGACCAGACGCAGGCTC | F:TTCCAGAGGAGGAGCATG R:ACTCAGCGTGACAACAGATAG | 14 |
Table 3 Specific information of off-target site
名称 Name | 脱靶序列 Off-target sequence | PCR引物序列 PCR primers’ sequence (5'-3') | 染色体位置 Chromosomal location |
|---|---|---|---|
| 脱靶-1 | GGACAGACAGAGGCGGGCTC | F:TGATGGATGTTGGCAGTGGT R:GCTGACAGACGCCGTGGTTC | 18 |
| 脱靶-2 | CCAGGACCAGACGCAGGCTC | F:TTCCAGAGGAGGAGCATG R:ACTCAGCGTGACAACAGATAG | 14 |

Fig. 2 Construction of sgRNA plasmidA: sgRNA-sg1 plasmid sequencing result; B: sgRNA-sg2 plasmid sequencing result; C: sgRNA-sg3 plasmid sequencing result
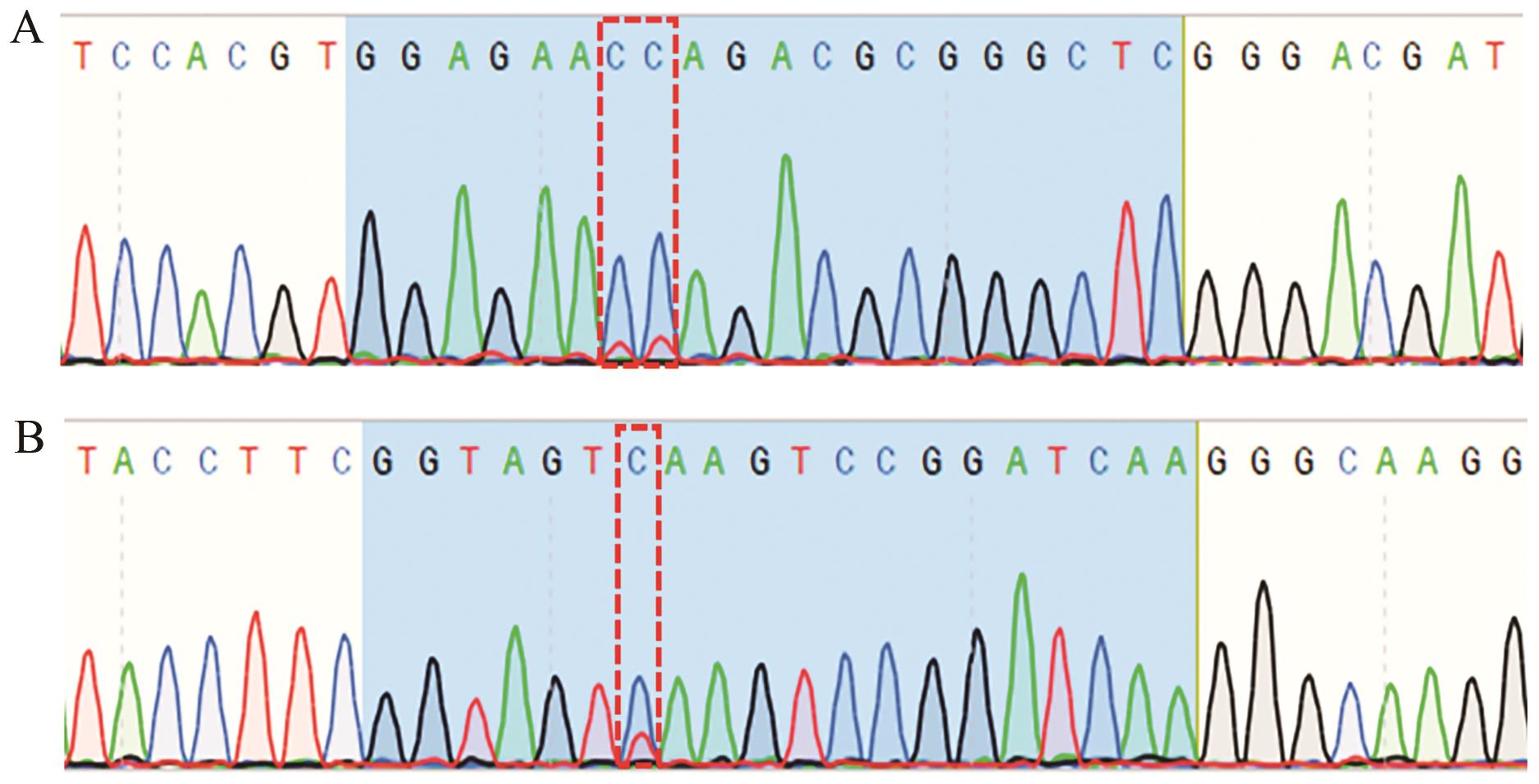
Fig. 4 Sequencing results of sgRNA-sg1 and sgRNA-sg3 PCR productsA: PCR sequencing results of sgRNA-sg1; B: PCR sequencing results of sgRNA-sg3. The base at the position of the red box is the site of mutation
sgRNA名称 sgRNA name | 靶位点序列信息 Target ID sequences (5′-3′) | 突变类型 Mutation | 突变比例 Mutation ratio (%) |
|---|---|---|---|
| sgRNA-sg1 | GGAGAACCAGACGCGGGCTCGGG GGAGAAT GGAGAATCAGACGCGGGCTCGGG GGAGAACCAGACGTGGGCTCGGG | WT C→T C→T C→T | 34.5(10/29) 13.8(4/29) 17.2(5/29) 3.4(1/29) |
| sgRNA-sg3 | GGTAGTCAAGTCCGGATCAAGGG GGTAGT GGTAG-------CGGATCAAGGG | WT C→T C→T,-7 bp/fs | 40.9(9/22) 36.4(8/22) 4.5(1/22) |
Table 4 T-A cloning results
sgRNA名称 sgRNA name | 靶位点序列信息 Target ID sequences (5′-3′) | 突变类型 Mutation | 突变比例 Mutation ratio (%) |
|---|---|---|---|
| sgRNA-sg1 | GGAGAACCAGACGCGGGCTCGGG GGAGAAT GGAGAATCAGACGCGGGCTCGGG GGAGAACCAGACGTGGGCTCGGG | WT C→T C→T C→T | 34.5(10/29) 13.8(4/29) 17.2(5/29) 3.4(1/29) |
| sgRNA-sg3 | GGTAGTCAAGTCCGGATCAAGGG GGTAGT GGTAG-------CGGATCAAGGG | WT C→T C→T,-7 bp/fs | 40.9(9/22) 36.4(8/22) 4.5(1/22) |
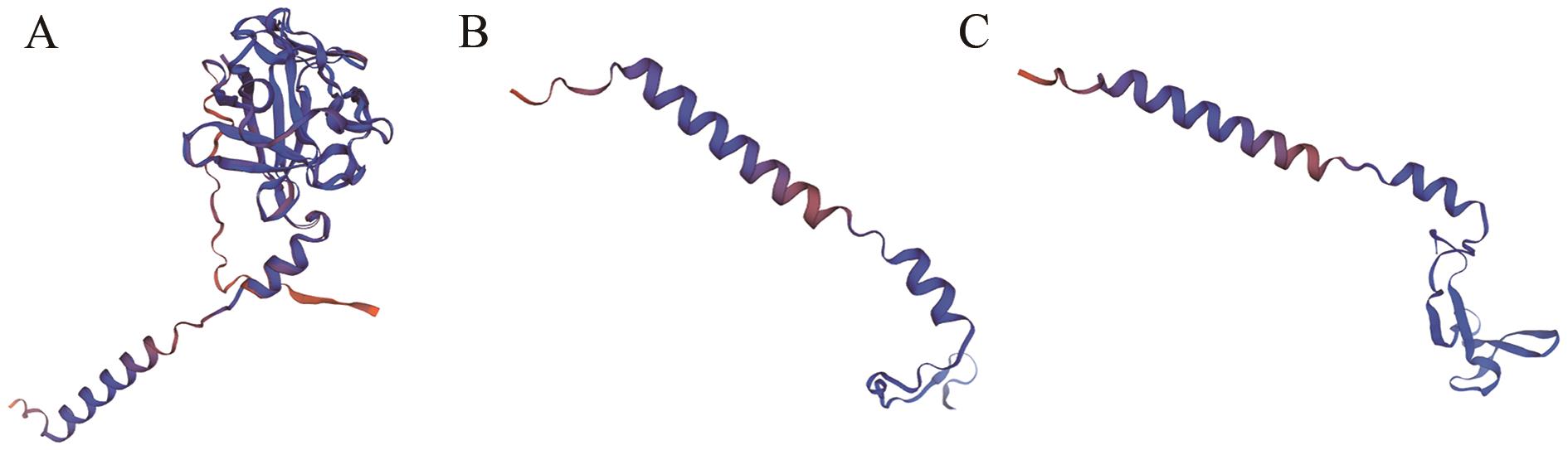
Fig. 5 Variation of protein structure of mutated FGF18 geneA: 3D model of wild-type-FGF18 protein; B: 3D model of mutant sgRNA-sg1 protein; C: 3D model of mutant sgRNA-sg3 protein
| [1] | Du YM, Liu YF, Hu JX, et al. CRISPR/Cas9 systems: Delivery technologies and biomedical applications [J]. Asian J Pharm Sci, 2023, 18(6): 100854. |
| [2] | Zong Y, Song QN, Li C, et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A [J]. Nat Biotechnol, 2018. |
| [3] | 王丽洁, 王潇, 杨力, 等. 碱基编辑技术的发展与应用 [J]. 生命的化学, 2019, 39(1): 13-20. |
| Wang LJ, Wang X, Yang L, et al. Development and application of base editor [J]. Chem Life, 2019, 39(1): 13-20. | |
| [4] | Zong Y, Wang YP, Li C, et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion [J]. Nat Biotechnol, 2017, 35(5): 438-440. |
| [5] | Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells [J]. Nat Rev Genet, 2018, 19(12): 770-788. |
| [6] | Li JY, Sun YW, Du JL, et al. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system [J]. Mol Plant, 2017, 10(3): 526-529. |
| [7] | Luo L, Shi Y, Wang HN, et al. Base editing in bovine embryos reveals a species-specific role of SOX2 in regulation of pluripotency [J]. PLoS Genet, 2022, 18(7): e1010307. |
| [8] | Shi Y, Hu BJ, Wang Z, et al. Functional role of GATA3 and CDX2 in lineage specification during bovine early embryonic development [J]. Reproduction, 2023, 165(3): 325-333. |
| [9] | Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway [J]. Wiley Interdiscip Rev Dev Biol, 2015, 4(3): 215-266. |
| [10] | Liu ZH, Lavine KJ, Hung IH, et al. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate [J]. Dev Biol, 2007, 302(1): 80-91. |
| [11] | Kawano M, Komi-Kuramochi A, Asada M, et al. Comprehensive analysis of FGF and FGFR expression in skin: FGF18 is highly expressed in hair follicles and capable of inducing anagen from telogen stage hair follicles [J]. J Investig Dermatol, 2005, 124(5): 877-885. |
| [12] | Kimura-Ueki M, Oda Y, Oki J, et al. Hair cycle resting phase is regulated by cyclic epithelial FGF18 signaling [J]. J Investig Dermatol, 2012, 132(5): 1338-1345. |
| [13] | Plikus MV. New activators and inhibitors in the hair cycle clock: targeting stem cells’ state of competence [J]. J Investig Dermatol, 2012, 132(5): 1321-1324. |
| [14] | Yu ZS, Ren MD, Wang ZX, et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila [J]. Genetics, 2013, 195(1): 289-291. |
| [15] | Barrangou R . events RNA. Cas9 targeting and the CRISPR revolution [J]. Science, 2014, 344(6185): 707-708. |
| [16] | 潘东霞, 王辉, 熊本海, 等. CRISPR-Cas基因编辑技术在羊生产应用中研究进展 [J]. 遗传, 2024, 46(9): 690-700. |
| Pan DX, Wang H, Xiong BH, et al. Progress on CRISPR-Cas gene editing technology in sheep production [J]. Hereditas: Beijing, 2024, 46(9): 690-700. | |
| [17] | Zheng W, Gu F. Progress of application and off-target effects of CRISPR/Cas9 [J]. Yi Chuan, 2015, 37(10): 1003-1010. |
| [18] | Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems [J]. Science, 2013, 339(6121): 819-823. |
| [19] | Mianné J, Chessum L, Kumar S, et al. Correction of the auditory phenotype in C57BL/6N mice via CRISPR/Cas9-mediated homology directed repair [J]. Genome Med, 2016, 8(1): 16. |
| [20] | Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage [J]. Nature, 2016, 533(7603): 420-424. |
| [21] | Xie JK, Ge WK, Li N, et al. Efficient base editing for multiple genes and loci in pigs using base editors [J]. Nat Commun, 2019, 10(1): 2852. |
| [22] | Lei LQ, Chen HQ, Xue W, et al. APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks [J]. Nat Struct Mol Biol, 2018, 25(1): 45-52. |
| [23] | Kim K, Ryu SM, Kim ST, et al. Highly efficient RNA-guided base editing in mouse embryos [J]. Nat Biotechnol, 2017, 35(5): 435-437. |
| [24] | Niu YY, Zhao XE, Zhou JK, et al. Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9 [J]. Reprod Fertil Dev, 2018, 30(2): 307-312. |
| [25] | Dang L, Li GL, Wang XJ, et al. Comparison of gene disruption induced by cytosine base editing-mediated iSTOP with CRISPR/Cas9-mediated frameshift [J]. Cell Prolif, 2020, 53(5): e12820. |
| [26] | 钟静丽, 林健香, 周建奎, 等. 碱基编辑系统的研究进展 [J]. 生物工程学报,2024,40(5):1271-1292. |
| Zhong JL, Lin JX, Zhou JK, et al. Research progress on base editing systems [J]. Chinese Journal of Biotechnology, 2024, 40(5): 1271-1292. | |
| [27] | Lee HK, Smith HE, Liu CY, et al. Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos [J]. Commun Biol, 2020, 3(1): 19. |
| [28] | Thuronyi BW, Koblan LW, Levy JM, et al. Continuous evolution of base editors with expanded target compatibility and improved activity [J]. Nat Biotechnol, 2019, 37(9): 1070-1079. |
| [29] | Li GW, Zhou SW, Li C, et al. Base pair editing in goat: nonsense codon introgression into FGF5 results in longer hair [J]. FEBS J, 2019, 286(23): 4675-4692. |
| [30] | Liu ZQ, Chen M, Chen SY, et al. Highly efficient RNA-guided base editing in rabbit [J]. Nat Commun, 2018, 9(1): 2717. |
| [31] | Molla KA, Yang YN. CRISPR/cas-mediated base editing: technical considerations and practical applications [J]. Trends Biotechnol, 2019, 37(10): 1121-1142. |
| [32] | 周勤, 王爽, 张婷, 等. 小鼠及猕猴胚胎MECP2基因T158M单碱基突变体系的建立 [J]. 中国生物工程杂志, 2020, 40(6):31-39. |
| Zhou Q, Wang S, Zhang T, et al. Establishment of a single-base mutation system for the T158M site in the MECP2 gene in mouse and macaque embryos [J]. China Biotechnology, 2020, 40(6): 31-39. | |
| [33] | Zhao JG, Lin HJ, Wang LS, et al. Suppression of FGF5 and FGF18 expression by cholesterol-modified siRNAs promotes hair growth in mice [J]. Front Pharmacol, 2021, 12: 666860. |
| [1] | DIAO Chen-yang, CUI You-zhi, LI Bing-zhi. Research Advances in Targeted Mutagenesis-mediated Microbial Evolutionary Engineering [J]. Biotechnology Bulletin, 2025, 41(8): 11-21. |
| [2] | DENG Mei-bi, YAN Lang, ZHAN Zhi-tian, ZHU Min, HE Yu-bing. Efficient CRISPR Gene Editing in Rice Assisted by RUBY [J]. Biotechnology Bulletin, 2025, 41(8): 65-73. |
| [3] | ZHOU Qian, TANG Meng-jun, ZHANG Xiao-yan, LU Jun-xian, TANG Xiu-jun, YANG Xing-xing, GAO Yu-shi. Research Progress in the Control of Multidrug Resistant Bacteria Based on in CRISPR-Cas System [J]. Biotechnology Bulletin, 2025, 41(5): 42-51. |
| [4] | CHEN Xiao-jun, HUI Jian, MA Hong-wen, BAI Hai-Bo, ZHONG Nan, LI Jia-run, FAN Yun-fang. Creating Rice Gerplasm Resources OsALS Rsistant to Herbicide through Single Base Gene Editing Technology [J]. Biotechnology Bulletin, 2025, 41(4): 106-114. |
| [5] | WEN Bo-lin, WAN Min, HU Jian-jun, WANG Ke-xiu, JING Sheng-lin, WANG Xin-yue, ZHU Bo, TANG Ming-xia, LI Bing, HE Wei, ZENG Zi-xian. Establishment of Genetic Transformation and Gene Editing System for a Potato Cultivar Chuanyu 50 [J]. Biotechnology Bulletin, 2025, 41(4): 88-97. |
| [6] | LIANG Li-cun, WANG Ke-fen, SONG Zu-huan, LIU Meng-ting, LI Jia-yu, LUO Hui-ying, YAO Bin, YANG Hao-meng. Improving the Efficiency of Gene Editing by Optimizing sgRNA in Aspergillus tubingensis [J]. Biotechnology Bulletin, 2025, 41(3): 62-70. |
| [7] | XUE Rui-ying, LIU Yong-ju, JIANG Yan-yan, PENG Xin-ya, CAO Dong, LI Yun, LIU Bao-long, BAO Xue-mei. Reducing the Expression of GBSSI Gene in Barley via the Editing in the 5′UTR Region [J]. Biotechnology Bulletin, 2025, 41(3): 83-89. |
| [8] | SUN Jing, YANG Yun-long, LIU Rong-zhi, YU Hong, LU Tie-gang. Strengthening Fundamental Research on Photosynthesis to Support Crop Breeding for High Yield [J]. Biotechnology Bulletin, 2025, 41(10): 1-5. |
| [9] | TONG Wei-jing, LUO Shu, LU Xin-lu, SHEN Jian-fu, LU Bai-yi, LI Kai-mian, MA Qiu-xiang, ZHANG Peng. CRISPR/Cas9 Editing MeHNL Gene to Generate Cassava Plants with Low Cyanogenic Glycoside [J]. Biotechnology Bulletin, 2024, 40(9): 11-19. |
| [10] | HOU Wen-ting, SUN Lin, ZHANG Yan-jun, DONG He-zhong. Application of Gene-editing Technology for Germplasm Innovation and Genetic Improvement in Cotton [J]. Biotechnology Bulletin, 2024, 40(7): 68-77. |
| [11] | LONG Jing, CHEN Jing-min, LIU Xiao, ZHANG Yi-fan, ZHOU Li-bin, DU Yan. Repair Mechanisms of DNA Double-strand Breaks and Their Roles in Heavy Ion Mutagenesis and Gene Editing in Plants [J]. Biotechnology Bulletin, 2024, 40(7): 55-67. |
| [12] | ZHOU Jia-wei, WU Zhi-qiang. Construction Method of mitoTALENs Mitochondrial Gene Editing Vector in Plants [J]. Biotechnology Bulletin, 2024, 40(10): 172-180. |
| [13] | LI Ming-kun, BI Mei-ying, ZHANG Tian-hang, WU Xiang-yu, YANG Pei-ru, YING Ming. Restoration of Agricultural Function of Rhizobacteria by UgRNA/Cas9 Multi-gene Editing [J]. Biotechnology Bulletin, 2024, 40(10): 275-287. |
| [14] | ZHANG Shuo, KAN Jun-hu, ZHOU Jia-wei, WU Zhi-qiang. Advance in Plant Mitochondrial Genome Editing [J]. Biotechnology Bulletin, 2024, 40(10): 41-52. |
| [15] | YANG Shuai-peng, QU Zi-xiao, ZHU Xiang-xing, TANG Dong-sheng. Optimization of DNA Base Editing Technology and Its Application in Pig Genetic Modification [J]. Biotechnology Bulletin, 2024, 40(1): 127-144. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||