








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (4): 190-201.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0966
Previous Articles Next Articles
YANG Ting( ), YANG Zong-tao, AI Jing, WANG Yu-tong, LI Yan-ye, DENG jun, LIU Jia-yong, ZHAO Yong(
), YANG Zong-tao, AI Jing, WANG Yu-tong, LI Yan-ye, DENG jun, LIU Jia-yong, ZHAO Yong( ), ZHANG Yue-bin
), ZHANG Yue-bin
Received:2025-09-09
Online:2026-02-09
Published:2026-02-09
Contact:
ZHAO Yong
E-mail:1146824026@qq.com;18087395132@163.com
YANG Ting, YANG Zong-tao, AI Jing, WANG Yu-tong, LI Yan-ye, DENG jun, LIU Jia-yong, ZHAO Yong, ZHANG Yue-bin. Analysis of Phenotypic Characteristics and Root Transcriptomics of Sugarcane with Different Genotypes[J]. Biotechnology Bulletin, 2026, 42(4): 190-201.
材料序号 Material number | 材料编码 Material code | 材料类型 Material type | 亲本信息(母本×父本) Parental cross (female × male) |
|---|---|---|---|
| 01 | 越南2号 | 野生种 | - |
| 02 | 云南82-1 | 野生种 | - |
| 03 | 2017-12-165 | 野生种 | - |
| 04 | 2017-22 | 野生种 | - |
| 05 | 云蔗0551 | 商品种 | - |
| 06 | A1 | 杂交种 | 云南82-1×2017-22 |
| 07 | A2 | 杂交种 | 云南82-1×2017-22 |
| 08 | B1 | 杂交种 | 越南2号×2017-12-165 |
| 09 | B2 | 杂交种 | 越南2号×2017-12-165 |
| 10 | N1 | 杂交种 | 云蔗0551×2017-22 |
| 11 | N2 | 杂交种 | 云蔗0551×越南2号 |
| 12 | N3 | 杂交种 | 云蔗0551×A1 |
| 13 | N4 | 杂交种 | 云蔗0551×B1 |
| 14 | N5 | 杂交种 | 云蔗0551×AB6 |
| 15 | N6 | 杂交种 | 云蔗0551×AB8 |
| 16 | AB6 | 杂交种 | A1×B1 |
| 17 | AB8 | 杂交种 | A1×B1 |
Table 1 Genetic backgrounds of 17 sugarcane materials
材料序号 Material number | 材料编码 Material code | 材料类型 Material type | 亲本信息(母本×父本) Parental cross (female × male) |
|---|---|---|---|
| 01 | 越南2号 | 野生种 | - |
| 02 | 云南82-1 | 野生种 | - |
| 03 | 2017-12-165 | 野生种 | - |
| 04 | 2017-22 | 野生种 | - |
| 05 | 云蔗0551 | 商品种 | - |
| 06 | A1 | 杂交种 | 云南82-1×2017-22 |
| 07 | A2 | 杂交种 | 云南82-1×2017-22 |
| 08 | B1 | 杂交种 | 越南2号×2017-12-165 |
| 09 | B2 | 杂交种 | 越南2号×2017-12-165 |
| 10 | N1 | 杂交种 | 云蔗0551×2017-22 |
| 11 | N2 | 杂交种 | 云蔗0551×越南2号 |
| 12 | N3 | 杂交种 | 云蔗0551×A1 |
| 13 | N4 | 杂交种 | 云蔗0551×B1 |
| 14 | N5 | 杂交种 | 云蔗0551×AB6 |
| 15 | N6 | 杂交种 | 云蔗0551×AB8 |
| 16 | AB6 | 杂交种 | A1×B1 |
| 17 | AB8 | 杂交种 | A1×B1 |
基因ID/内参名称 Gene ID/Internal reference name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) |
|---|---|---|
| Sspon.02G0008140-1T | GCCCATGGTGGTGGAGAC | CAACAGCCACCGTTTGCC |
| Sspon.05G0002890-3D | TTACCAGTGGGCTTGCGG | GCAAGTGCAGCCACATCG |
| Sspon.02G0013210-1A | ACTCGTCACAGCCAACCG | CCCGTCGCGTCGAATACA |
| Sspon.07G0022280-2C | GCCAGCGACTCTGCATCT | TGCTGCTGTGCAAAAGGC |
| Sspon.04G0031010-1P | GAGTGCCCTGTGTCGCAT | GTCTGGCCGCTGAACCTT |
| Sspon.01G0014700-2B | GGCTCCGAGGTTGCAGTT | TCTGAGCTTCTGCCCCCA |
| 25S rRNA1 | CCTGAAGATCACCCTGTGCT | GCAGTCTCCAGCTCCTGTTC |
Table 2 Primers used for RT-qPCR
基因ID/内参名称 Gene ID/Internal reference name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) |
|---|---|---|
| Sspon.02G0008140-1T | GCCCATGGTGGTGGAGAC | CAACAGCCACCGTTTGCC |
| Sspon.05G0002890-3D | TTACCAGTGGGCTTGCGG | GCAAGTGCAGCCACATCG |
| Sspon.02G0013210-1A | ACTCGTCACAGCCAACCG | CCCGTCGCGTCGAATACA |
| Sspon.07G0022280-2C | GCCAGCGACTCTGCATCT | TGCTGCTGTGCAAAAGGC |
| Sspon.04G0031010-1P | GAGTGCCCTGTGTCGCAT | GTCTGGCCGCTGAACCTT |
| Sspon.01G0014700-2B | GGCTCCGAGGTTGCAGTT | TCTGAGCTTCTGCCCCCA |
| 25S rRNA1 | CCTGAAGATCACCCTGTGCT | GCAGTCTCCAGCTCCTGTTC |
性状 Trait | 最小值 Min | 最大值 Max | 平均值 Average | 极差 Range | 标准差 S | 变异系数 CV | 遗传多样性指数 H′ |
|---|---|---|---|---|---|---|---|
| 株高 PH (cm) | 218.00 | 314.60 | 253.61 | 96.60 | 24.67 | 0.10 | 2.83 |
| 茎径 SD (cm) | 0.74 | 2.51 | 1.28 | 1.77 | 0.48 | 0.37 | 2.77 |
| 茎节数 SNN (Nodes/m) | 16.40 | 31.00 | 24.05 | 14.60 | 3.59 | 0.15 | 2.82 |
| 节间长度 STL (cm) | 18.10 | 29.84 | 22.86 | 11.74 | 3.20 | 0.14 | 2.82 |
| 单茎重 SNW (kg) | 0.09 | 1.26 | 0.36 | 1.17 | 0.31 | 0.84 | 2.55 |
| 有效茎数 ESN (Stem nodes/plant) | 5.80 | 44.80 | 20.40 | 39.00 | 12.34 | 0.61 | 2.66 |
Table 3 Basic statistical analysis of 7 agronomic traits of 17 sugarcane materials
性状 Trait | 最小值 Min | 最大值 Max | 平均值 Average | 极差 Range | 标准差 S | 变异系数 CV | 遗传多样性指数 H′ |
|---|---|---|---|---|---|---|---|
| 株高 PH (cm) | 218.00 | 314.60 | 253.61 | 96.60 | 24.67 | 0.10 | 2.83 |
| 茎径 SD (cm) | 0.74 | 2.51 | 1.28 | 1.77 | 0.48 | 0.37 | 2.77 |
| 茎节数 SNN (Nodes/m) | 16.40 | 31.00 | 24.05 | 14.60 | 3.59 | 0.15 | 2.82 |
| 节间长度 STL (cm) | 18.10 | 29.84 | 22.86 | 11.74 | 3.20 | 0.14 | 2.82 |
| 单茎重 SNW (kg) | 0.09 | 1.26 | 0.36 | 1.17 | 0.31 | 0.84 | 2.55 |
| 有效茎数 ESN (Stem nodes/plant) | 5.80 | 44.80 | 20.40 | 39.00 | 12.34 | 0.61 | 2.66 |
性状 Trait | 项目 Item | 种质类群 Germplasm population | ||
|---|---|---|---|---|
| Group Ι | Group Ⅱ | |||
| 株高PH (cm) | 平均值 Average | 252.57 | 254.34 | |
| 变异系数 CV | 0.09 | 0.11 | ||
| 平均茎径ASD (cm) | 平均值 Average | 1.12 | 1.39 | |
| 变异系数 CV | 0.56 | 0.24 | ||
| 茎节数SNN (Nodes/m) | 平均值 Average | 21.91 | 25.54 | |
| 变异系数 CV | 0.18 | 0.10 | ||
| 节间长度STL (cm) | 平均值 Average | 22.87 | 22.85 | |
| 变异系数 CV | 0.15 | 0.14 | ||
| 单茎重SNW (kg) | 平均值 Average | 0.31 | 0.40 | |
| 变异系数 CV | 1.39 | 0.51 | ||
| 有效茎数ESN(Stem nodes/plant) | 平均值 Average | 28.00 | 15.08 | |
| 变异系数 CV | 0.42 | 0.67 | ||
| 锤度B | 平均值 Average | 11.10 | 14.94 | |
| 变异系数 CV | 0.41 | 0.33 | ||
Table 4 Comparison of agronomic traits between two groups of sugarcane materials
性状 Trait | 项目 Item | 种质类群 Germplasm population | ||
|---|---|---|---|---|
| Group Ι | Group Ⅱ | |||
| 株高PH (cm) | 平均值 Average | 252.57 | 254.34 | |
| 变异系数 CV | 0.09 | 0.11 | ||
| 平均茎径ASD (cm) | 平均值 Average | 1.12 | 1.39 | |
| 变异系数 CV | 0.56 | 0.24 | ||
| 茎节数SNN (Nodes/m) | 平均值 Average | 21.91 | 25.54 | |
| 变异系数 CV | 0.18 | 0.10 | ||
| 节间长度STL (cm) | 平均值 Average | 22.87 | 22.85 | |
| 变异系数 CV | 0.15 | 0.14 | ||
| 单茎重SNW (kg) | 平均值 Average | 0.31 | 0.40 | |
| 变异系数 CV | 1.39 | 0.51 | ||
| 有效茎数ESN(Stem nodes/plant) | 平均值 Average | 28.00 | 15.08 | |
| 变异系数 CV | 0.42 | 0.67 | ||
| 锤度B | 平均值 Average | 11.10 | 14.94 | |
| 变异系数 CV | 0.41 | 0.33 | ||
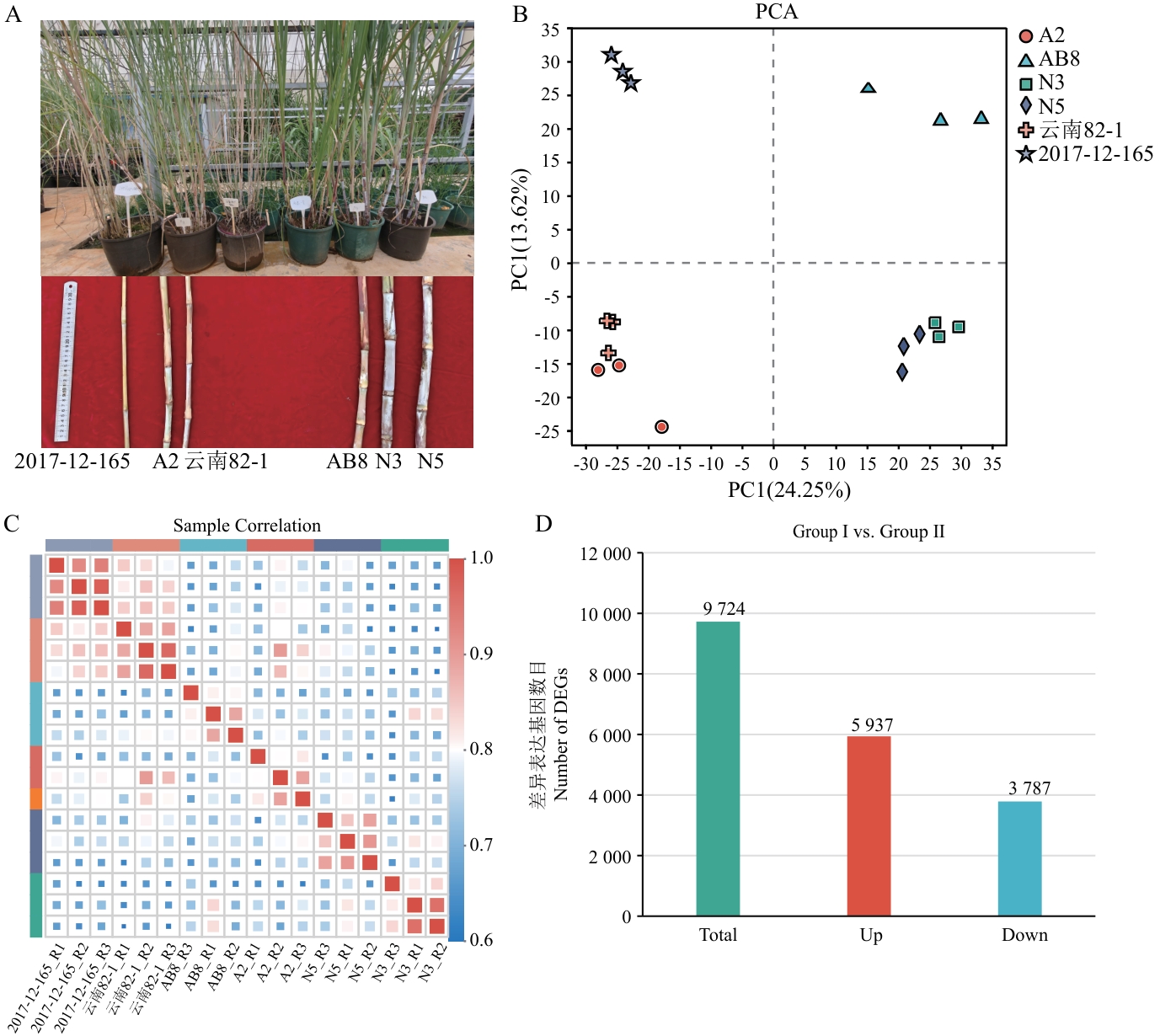
Fig. 2 Transcriptome sequencing analysisA: Phenotypes of six materials in two groups. B: PCA analysis. C: Correlation analysis among samples. D: Bar chart of DEGs between Group Ι and Group Ⅱ
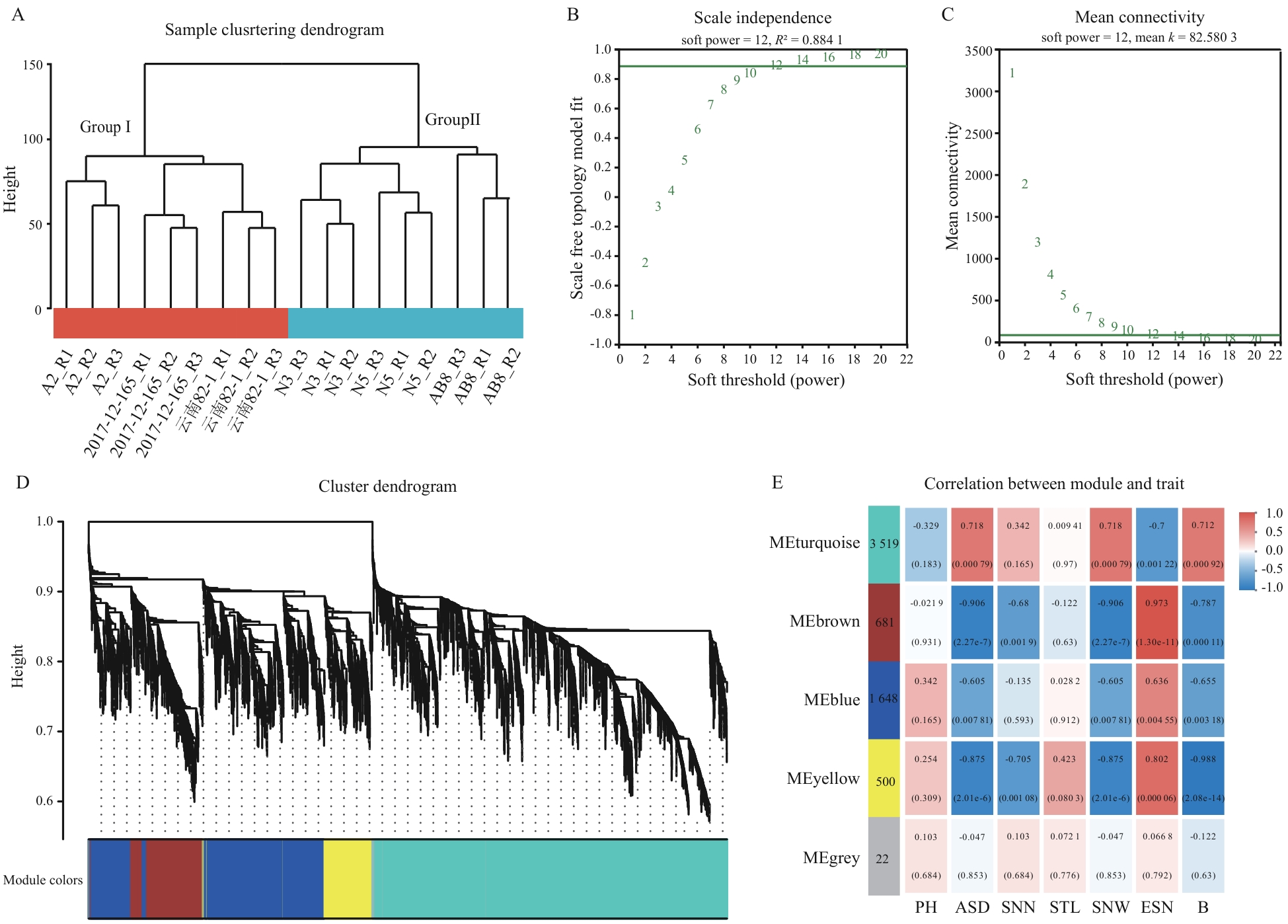
Fig. 4 Construction of WGCNAA: Sample clustering. B: Scale-free fit curve and average connectivity curve. C: Gene clustering and module construction. D: Heatmap of the correlation between modules and phenotypes (red grids indicate positive correlations between samples and modules, and purple grids indicate negative correlations between samples and modules. The values inside and outside the parentheses in the figure are the P-value and the correlation coefficient r, respectively)
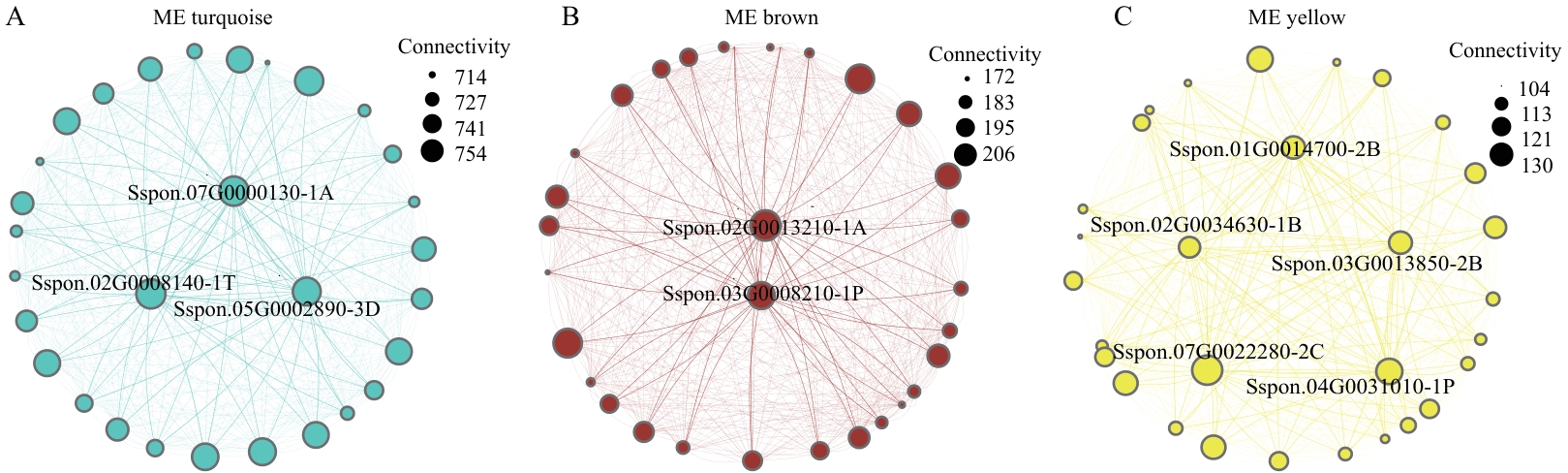
Fig. 5 Interaction networks of hub genes within key modulesA: Turquoise module. B: Brown module. C: Yellow module (The sizes of nodes and labels indicatethe connectivity of genes)
模块 Module | 基因号 Gene ID | 功能注释 Annotation | KEGG注释 KEGG pathway | 蛋白结构域 PFAMs |
|---|---|---|---|---|
蓝绿色 Turquoise | Sspon.07G0000130-1A | Protein kinase superfamily | MAPK signaling pathway | Pkinase |
| Sspon.02G0008140-1T | This protein promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis | RNA transport | GTP_EFTU | |
| Sspon.05G0002890-3D | SIT4 phosphatase-associated protein | - | SAPS | |
棕色 Brown | Sspon.02G0013210-1A | DNA-binding domain in plant proteins such as APETALA2 and EREBPs | - | AP2 |
| Sspon.03G0008210-1P | Protein kinase domain | MAPK signaling pathway | Pkinase | |
黄色 Yellow | Sspon.07G0022280-2C | Belongs to the syntaxin family | SNARE interactions in vesicular transport | Syntaxin |
| Sspon.04G0031010-1P | Phosphotransferase enzyme family | Phosphotransferase enzyme family | ||
| Sspon.01G0014700-2B | Acyl transferase domain | Fatty acid biosynthesis | Acyl_transf_1 | |
| Sspon.03G0013850-2B | Dienelactone hydrolase family | - | NUDIX | |
| Sspon.02G0034630-1B | Belongs to the glycosyl hydrolase 17 family | Starch and sucrose metabolism | Glyco_hydro_17 |
Table 5 Candidate hub genes identified by WGCNA and their functional annotations
模块 Module | 基因号 Gene ID | 功能注释 Annotation | KEGG注释 KEGG pathway | 蛋白结构域 PFAMs |
|---|---|---|---|---|
蓝绿色 Turquoise | Sspon.07G0000130-1A | Protein kinase superfamily | MAPK signaling pathway | Pkinase |
| Sspon.02G0008140-1T | This protein promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during protein biosynthesis | RNA transport | GTP_EFTU | |
| Sspon.05G0002890-3D | SIT4 phosphatase-associated protein | - | SAPS | |
棕色 Brown | Sspon.02G0013210-1A | DNA-binding domain in plant proteins such as APETALA2 and EREBPs | - | AP2 |
| Sspon.03G0008210-1P | Protein kinase domain | MAPK signaling pathway | Pkinase | |
黄色 Yellow | Sspon.07G0022280-2C | Belongs to the syntaxin family | SNARE interactions in vesicular transport | Syntaxin |
| Sspon.04G0031010-1P | Phosphotransferase enzyme family | Phosphotransferase enzyme family | ||
| Sspon.01G0014700-2B | Acyl transferase domain | Fatty acid biosynthesis | Acyl_transf_1 | |
| Sspon.03G0013850-2B | Dienelactone hydrolase family | - | NUDIX | |
| Sspon.02G0034630-1B | Belongs to the glycosyl hydrolase 17 family | Starch and sucrose metabolism | Glyco_hydro_17 |
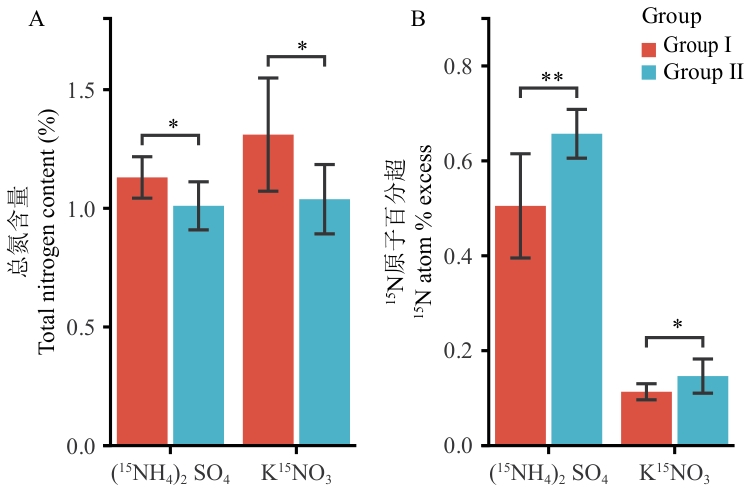
Fig. 7 Dynamics of root total nitrogen content (A) and ¹⁵N uptake (B) under ammonium and nitrate nutrition in two sugarcane groupsData are presented as mean ± SD (n = 3). Significant differences between Group I and Group II under the same nitrogen source were determined by independent-samples t test. * and ** indicate significant differences at P < 0.05 and P < 0.01, respectively
| [1] | Islam MS, Yang XP, Sood S, et al. Molecular characterization of genetic basis of Sugarcane yellow leaf virus (SCYLV) resistance in Saccharum spp. hybrid [J]. Plant Breed, 2018, 137(4): 598-604. |
| [2] | Sharma A, Chandra A. Identification of new Leuconostoc species responsible for post-harvest sucrose losses in sugarcane [J]. Sugar Tech, 2018, 20(4): 492-496. |
| [3] | Goldemberg J. The Brazilian biofuels industry [J]. Biotechnol Biofuels, 2008, 1(1): 6. |
| [4] | Frink CR, Waggoner PE, Ausubel JH. Nitrogen fertilizer: retrospect and prospect [J]. Proc Natl Acad Sci USA, 1999, 96(4): 1175-1180. |
| [5] | Garnett T, Conn V, Kaiser BN. Root based approaches to improving nitrogen use efficiency in plants [J]. Plant Cell Environ, 2009, 32(9): 1272-1283. |
| [6] | 翁梦静, 张慈凤, 林娜平, 等. 甘蔗ScmiR393在植物应答缺氮胁迫中的功能研究 [J]. 福建农业科技, 2025, 56(1): 24-30. |
| Weng MJ, Zhang CF, Lin NP, et al. Functional study of sugarcane ScmiR393 in plant response to nitrogen deficiency stress [J]. Fujian Agric Sci Technol, 2025, 56(1): 24-30. | |
| [7] | Robinson N, Brackin R, Vinall K, et al. Nitrate paradigm does not hold up for sugarcane [J]. PLoS One, 2011, 6(4): e19045. |
| [8] | 李文宝. 甘蔗营养高效利用种质资源筛选研究 [D]. 南宁: 广西大学, 2011. |
| Li WB. Screenning for higher nutrients use efficiency of sugarcane germplasm [D]. Nanning: Guangxi University, 2011. | |
| [9] | 杨荣仲, 周会, 桂意云, 等. 甘蔗低氮胁迫性状变化与蔗茎含氮量的研究 [J]. 安徽农业科学, 2013, 41(2): 481-484. |
| Yang RZ, Zhou H, Gui YY, et al. Response to low nitrogen stress and stalk nitrogen content of sugarcane germplasm [J]. J Anhui Agric Sci, 2013, 41(2): 481-484. | |
| [10] | Fan XR, Feng HM, Tan YW, et al. A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen [J]. J Integr Plant Biol, 2016, 58(6): 590-599. |
| [11] | Ge M, Wang YC, Liu YH, et al. The NIN-like protein 5 (ZmNLP5) transcription factor is involved in modulating the nitrogen response in maize [J]. Plant J, 2020, 102(2): 353-368. |
| [12] | 杨文亭, 李志贤, 赖健宁, 等. 甘蔗‒大豆间作和减量施氮对甘蔗产量和主要农艺性状的影响 [J]. 作物学报, 2014, 40(3): 556-562. |
| Yang WT, Li ZX, Lai JN, et al. Effects of sugarcane-soybean intercropping and reduced nitrogen application on yield and major agronomic traits of sugarcane [J]. Acta Agron Sin, 2014, 40(3): 556-562. | |
| [13] | de Castro SGQ, Magalhães PSG, de Castro SAQ, et al. Optimizing nitrogen fertilizer rates at distinct in-season application moments in sugarcane [J]. Int J Plant Prod, 2022, 16(1): 137-152. |
| [14] | 张艳梅. 施氮水平对不同甘蔗品种产量、品质及生理生化特性的影响 [D]. 南宁: 广西大学, 2015. |
| Zhang YM. Effects of nitrogen levels on cane yield and quality, and physiological and biochemical characteristics of different sugarcane varieties [D]. Nanning: Guangxi University, 2015. | |
| [15] | Huang DL, Qin CX, Gui YY, et al. Role of the SPS gene families in the regulation of sucrose accumulation in sugarcane [J]. Sugar Tech, 2017, 19(2): 117-124. |
| [16] | Verma AK, Upadhyay SK, Verma PC, et al. Functional analysis of sucrose phosphate synthase (SPS) and sucrose synthase (SS) in sugarcane (Saccharum) cultivars [J]. Plant Biol, 2011, 13(2): 325-332. |
| [17] | 陈俊华, 文吉富, 王国良, 等. Excel在计算群落生物多样性指数中的应用 [J]. 四川林业科技, 2009, 30(3): 88-90, 60. |
| Chen JH, Wen JF, Wang GL, et al. On application of excel in calculating the biodiversity index of communities [J]. J Sichuan For Sci Technol, 2009, 30(3): 88-90, 60. | |
| [18] | 李旭娟, 李纯佳, 刘洪博, 等. 甘蔗腋芽形成发育过程的转录组分析 [J]. 生物技术通报, 2025, 41(3): 202-218. |
| Li XJ, Li CJ, Liu HB, et al. Transcriptome analysis of axillary bud formation and development in sugarcane [J]. Biotechnol Bull, 2025, 41(3): 202-218. | |
| [19] | 阙友雄, 许莉萍, 徐景升, 等. 甘蔗基因表达定量PCR分析中内参基因的选择 [J]. 热带作物学报, 2009, 30(3): 274-278. |
| Que YX, Xu LP, Xu JS, et al. Selection of control genes in real-time qPCR analysis of gene expression in sugarcane [J]. Chin J Trop Crops, 2009, 30(3): 274-278. | |
| [20] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [21] | 李俊杰, 杜蒲芳, 石婷瑞, 等. 不同基因型小麦苗期耐低氮性评价及筛选 [J]. 中国农业科技导报, 2021, 23(7): 21-32. |
| Li JJ, Du PF, Shi TR, et al. Screening and evaluation of low nitrogen tolerance from different genotypes wheat at seedling stage [J]. J Agric Sci Technol, 2021, 23(7): 21-32. | |
| [22] | Liu YQ, Wang HR, Jiang ZM, et al. Genomic basis of geographical adaptation to soil nitrogen in rice [J]. Nature, 2021, 590(7847): 600-605. |
| [23] | Zhan J, Zhou YF, Yang LS, et al. Low ammonium and high nitrate input improves nitrogen use efficiency and growth in sugarcane through coordinated reprogramming of nitrogen and carbon metabolism [J]. Plant Physiol Biochem, 2025, 229: 110354. |
| [24] | 江厚龙, 李勇, 陈天才, 等. 综合转录组和代谢组分析揭示嫁接提高烟草钾利用效率的分子机制 [J]. 天津农业科学, 2025, 31(4): 1-12, 19. |
| Jiang HL, Li Y, Chen TC, et al. Integrated transcriptomic and metabolomic analyses elucidate the mechanism underlying the impact of grafting on potassium utilization efficiency in tobacco [J]. Tianjin Agric Sci, 2025, 31(4): 1-12, 19. | |
| [25] | 李寒雪. 代谢组和转录组联合解析藜麦幼苗对氮素的响应机制 [D]. 昆明: 云南农业大学, 2024. |
| Li HX. Transcriptome and metabolome analyses revealed the response mechanism of quinoa seedlings to nitrogen fertilizer [D]. Kunming: Yunnan Agricultural University, 2024. | |
| [26] | 南运有. BnaA5.AIB与BnaC 2.NAR2.1参与甘蓝型油菜对外源脱落酸的应答响应及氮吸收利用的调控 [D]. 杨凌: 西北农林科技大学, 2023. |
| Nan YY. BnaA5.AIB and BnaC 2.NAR2.1 are involved in the response to exogenous abscisic acid and the regulation of nitrogen uptake and utilization in Brassica napus L. [D]. Yangling: Northwest A & F University, 2023. | |
| [27] | 葛礼姣. 氮高效利用菊花品种筛选及高效利用机理研究 [D]. 南京: 南京农业大学, 2022. |
| Ge LJ. Study on nitrogen efficient variety screening and its mechanism on high nitrogen utilization efficiency in Chrysanthemum [D]. Nanjing: Nanjing Agricultural University, 2022. | |
| [28] | 任盼荣, 汪军成, 姚立蓉, 等. 大麦种质资源磷利用效率的评价及其转录组分析 [J]. 大麦与谷类科学, 2018, 35(4): 56-57. |
| Ren PR, Wang JC, Yao LR, et al. Evaluation of phosphorus use efficiency and transcriptome analysis in barley germplasm (Hordeum vulgare L.) [J]. Barley Cereal Sci, 2018, 35(4): 56-57. | |
| [29] | Liu H, Gao XH, Fan WS, et al. Optimizing carbon and nitrogen metabolism in plants: From fundamental principles to practical applications [J]. J Integr Plant Biol, 2025, 67(6): 1447-1466. |
| [30] | 刘潇潇. 膜脂在小麦低氮胁迫响应中的作用及机制研究 [D]. 北京: 中国科学院大学(中国科学院教育部水土保持与生态环境研究中心), 2021. |
| Liu XX. The mechanism of the involvement of leaf membrane lipids in response to nitrogen deficiency in wheat [D]. Beijing: Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, 2021. | |
| [31] | 梁桂红, 华营鹏, 周婷, 等. 甘蓝型油菜NRT1.5和NRT1.8家族基因的生物信息学分析及其对氮-镉胁迫的响应 [J]. 作物学报, 2019, 45(3): 365-380. |
| Liang GH, Hua YP, Zhou T, et al. Bioinformatics analysis and response to nitrate-cadmium stress of NRT1.5 and NRT1.8 family genes in Brassica napus [J]. Acta Agron Sin, 2019, 45(3): 365-380. | |
| [32] | 钱甫, 张占琴, 陈树宾, 等. 基于GWAS和WGCNA分析挖掘玉米花期相关候选基因 [J]. 作物学报, 2023, 49(12): 3261-3276. |
| Qian F, Zhang ZQ, Chen SB, et al. Mining maize flowering traits related candidate genes based on GWAS and WGCNA data [J]. Acta Agron Sin, 2023, 49(12): 3261-3276. | |
| [33] | 郝瑞杰, 邱晨, 耿晓云, 等. 梅花PmABCG9在苯甲醇挥发中的功能分析 [J]. 中国农业科学, 2023, 56(13): 2574-2585. |
| Hao RJ, Qiu C, Geng XY, et al. The function of PmABCG9 transporter related to the volatilization of benzyl alcohol in Prunus mume [J]. Sci Agric Sin, 2023, 56(13): 2574-2585. | |
| [34] | 荐红举, 张梅花, 尚丽娜, 等. 利用WGCNA筛选马铃薯块茎发育候选基因 [J]. 作物学报, 2022, 48(7): 1658-1668. |
| Jian HJ, Zhang MH, Shang LN, et al. Screening candidate genes involved in potato tuber development using WGCNA [J]. Acta Agron Sin, 2022, 48(7): 1658-1668. | |
| [35] | 张恒燕, 张木清. 基于WGCNA挖掘外源GA3调节甘蔗节间伸长相关基因 [J/OL]. 分子植物育种, 2022. . |
| Zhang HY, Zhang MQ. Mining of genes related to exogenous GA3 regulation of sugarcane internode elongation based on WGCNA [J/OL]. Mol Plant Breed, 2022. . | |
| [36] | Wang L, Ming LC, Liao KY, et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice [J]. Mol Plant, 2021, 14(7): 1168-1184. |
| [37] | 汪宽鸿, 祝彪, 朱祝军. GSH/GSSG在植物应对非生物胁迫中的作用综述 [J]. 园艺学报, 2021, 48(4): 647-660. |
| Wang KH, Zhu B, Zhu ZJ. Review of the role of GSH/GSSG in plant abiotic stress response [J]. Acta Hortic Sin, 2021, 48(4): 647-660. | |
| [38] | 祁雪姣, 谢乾坤, 谢雨欣, 等. 籼稻品种Y两优886高氮素利用效率机制研究 [J]. 河南农业科学, 2021, 50(3): 25-32. |
| Qi XJ, Xie QK, Xie YX, et al. Mechanism of high nitrogen utilization efficiency of indica rice yliangyou 886 [J]. J Henan Agric Sci, 2021, 50(3): 25-32. | |
| [39] | 吕江艳, 龙鹏宇, 罗维钢, 等. 甘蔗节水高产和蔗田氧化亚氮减排的滴灌施肥模式 [J]. 节水灌溉, 2023(12): 1-8. |
| Lü JY, Long PY, Luo WG, et al. Drip irrigation fertilization model for water-saving and high-yield sugarcane and reduction of nitrous oxide emissions in sugarcane fields [J]. Water Sav Irrig, 2023(12): 1-8. |
| [1] | WANG Yu-kun, YUAN Yuan, WANG Bin, ZHU Yun-na, REN Xiao-qiang, REN Fei, YE Hong. Integrated Analysis of Transcriptome and Lipid Metabolome Reveals the Differences in α-Linolenic Acid Synthesis Regulation in Different Perilla frutescens [J]. Biotechnology Bulletin, 2026, 42(4): 129-140. |
| [2] | LIU Qing-yuan, WU Hong-qi, CHEN Xiu-e, CHEN Jian, JIANG Yuan-ze, HE Yan-zi, YU Qi-wei, LIU Ren-xiang. Function of Transcription Factor NtMYB96a in Regulating the Tolerance of Tobacco to Drought [J]. Biotechnology Bulletin, 2026, 42(4): 239-250. |
| [3] | FAN Rong-hui, LUO Yuan-hua, CHEN Yi-quan, FANG Neng-yan, CHEN Yan, ZHONG Huai-qin, YE Xiu-xian. Comparative Analysis of Floral Scent Formation between ‘Jinhui’ and Onc. ‘Sharry Baby’ of Oncidium hybridum [J]. Biotechnology Bulletin, 2026, 42(1): 105-113. |
| [4] | LIU Jian-guo, LIU Ge-er, GUO Ying-xin, WANG Bin, WANG Yu-kun, LU Jin-feng, HUANG Wen-ting, ZHU Yun-na. Integrate Transcriptomic and Metabolomic Analysis of Fruits Quality Differences between ‘Guiyou No. 1’ and ‘Shatianyou’ Pomelo (Citrus maxima) [J]. Biotechnology Bulletin, 2025, 41(9): 168-181. |
| [5] | LIU Ze-zhou, DUAN Nai-bin, YUE Li-xin, WANG Qing-hua, YAO Xing-hao, GAO Li-min, KONG Su-ping. Analysis of Wax Components and Screening of Wax-deficient Gene Ggl-1 in Garlic (Allium sativum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 219-231. |
| [6] | YAN Meng-yang, LIANG Xiao-yang, DAI Jun-ang, ZHANG Yan, GUAN Tuan, ZHANG Hui, LIU Liang-bo, SUN Zhi-hua. Screening of Amoxicillin-degrading Bacteria and Study on Its Degradation Mechanisms [J]. Biotechnology Bulletin, 2025, 41(9): 314-325. |
| [7] | ZHU Li-juan, ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li. Mining the Core Genes Being Tolerant to Cadmium in Wild Soybean by WGCNA [J]. Biotechnology Bulletin, 2025, 41(8): 124-136. |
| [8] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [9] | WANG Yue-chen, HAN Xin-qi, WEI Wen-min, CUI Zhao-lan, LUO Yang-mei, CHEN Peng-ru, WANG Hai-gang, LIU long-long, ZHANG Li, WANG Lun. Biological Basis Study for Grain Shattering in Proso Millet and Identification of Genes Regulating Grain Shattering [J]. Biotechnology Bulletin, 2025, 41(7): 164-171. |
| [10] | ZHANG Jin-hao, DENG Hui, ZHANG Qing-zhuang, TAO Yu, ZHOU Chi, LI Xin. Modulation of the Growth, Quality, and Cadmium Content of Lily Bulbs by Bacillus velezensis XY40-1 [J]. Biotechnology Bulletin, 2025, 41(7): 281-291. |
| [11] | ZHANG Yue, BI Yu, MU Xue-nan, ZHENG Zi-wei, WANG Zhi-gang, XU Wei-hui. Biocontrol Characteristics of Strain JB7 against Fusarium graminearum [J]. Biotechnology Bulletin, 2025, 41(7): 261-271. |
| [12] | DONG Xu-kun, CHE Yong-mei, WANG Ming-shuo, LUO Zheng-gang, GUAN En-sen, ZHAO Fang-gui, YE Qing, LIU Xin. Effects of Co-treatment of Nano-silica and Bacillus cereus SS1 on the Growth of Tobacco [J]. Biotechnology Bulletin, 2025, 41(7): 292-298. |
| [13] | LI Cheng-hua, DOU Fei-fei, REN Yu-zhao, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Effect of Exogenous Salicylic Acid on Wheat Infested with Blumeria graminis f. sp. tritici and Its Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(7): 272-280. |
| [14] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [15] | GUO Xiu-juan, FENG Yu, WU Rui-xiang, WANG Li-qin, YANG Jian-chun. Transcriptome Analysis of the Effect of Ca 2+ Treatment on the Seed Germination of Flax [J]. Biotechnology Bulletin, 2025, 41(7): 139-149. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||