








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (3): 278-289.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0732
Previous Articles Next Articles
XU Xiao-wen1( ), LI Jin-cang2, HAI Du2, ZHA Yu-ping1, SONG Fei1, WANG Yi-xun1(
), LI Jin-cang2, HAI Du2, ZHA Yu-ping1, SONG Fei1, WANG Yi-xun1( )
)
Received:2022-06-17
Online:2023-03-26
Published:2023-04-10
XU Xiao-wen, LI Jin-cang, HAI Du, ZHA Yu-ping, SONG Fei, WANG Yi-xun. Identification and Diversity Analysis of Mycoviruses from the Phytopathogenic Fungus Colletotrichum spp. of Walnut[J]. Biotechnology Bulletin, 2023, 39(3): 278-289.
| 种名Species | 菌株编号Strain No. | 采集地点Collecting location | 种名Species | 菌株编号Strain No. | 采集地点Collecting location | |
|---|---|---|---|---|---|---|
| C. gloeosporioides | SZ2-3 | Shangzhou, Shanxi, China | C. siamense | YX1-5 | Yunxi, Hubei, China | |
| C. gloeosporioides | DJ1-1 | Danjiang, Hubei, China | C. fioriniae | BK3-5 | Baokang, Hubei, China | |
| C. gloeosporioides | DJ2-1 | Danjiang, Hubei, China | C. fioriniae | CX3 | Chengxian, Gansu, China | |
| C. gloeosporioides | DJ2-4 | Danjiang, Hubei, China | C. fioriniae | BK2-3 | Baokang, Hubei, China | |
| C. gloeosporioides | YY1-11 | Yunyang, Hubei, China | C. fioriniae | CX1 | Chengxian, Gansu, China | |
| C. fructicola | XS6 | Xingshan, Hubei, China | C. fioriniae | CX7 | Chengxian, Gansu, China | |
| C. fructicola | FX1-3 | Fengxiang, Shanxi, China | C. nymphaeae | ZS3-13 | Zhushan, Hubei, China | |
| C. fructicola | CX6 | Chengxian, Gansu, China | C. nymphaeae | ZS3-8 | Zhushan, Hubei, China | |
| C. fructicola | YY1-28 | Yunyang, Hubei, China | C. nymphaeae | BK1-11 | Baokang, Hubei, China | |
| C. fructicola | BK2-1 | Baokang, Hubei, China | C. nymphaeae | ZS2-6 | Zhushan, Hubei, China | |
| C. siamense | YX1-7 | Yunxi, Hubei, China | C. nymphaeae | XS5 | Xingshan, Hubei, China | |
| C. siamense | ZS2-9 | Zhushan, Hubei, China | C. nymphaeae | BK4-4 | Baokang, Hubei, China | |
| C. siamense | DJ3-5 | Danjiang, Hubei, China |
Table 1 Information of the Colletotrichum species from 25 walnut plants for experiment
| 种名Species | 菌株编号Strain No. | 采集地点Collecting location | 种名Species | 菌株编号Strain No. | 采集地点Collecting location | |
|---|---|---|---|---|---|---|
| C. gloeosporioides | SZ2-3 | Shangzhou, Shanxi, China | C. siamense | YX1-5 | Yunxi, Hubei, China | |
| C. gloeosporioides | DJ1-1 | Danjiang, Hubei, China | C. fioriniae | BK3-5 | Baokang, Hubei, China | |
| C. gloeosporioides | DJ2-1 | Danjiang, Hubei, China | C. fioriniae | CX3 | Chengxian, Gansu, China | |
| C. gloeosporioides | DJ2-4 | Danjiang, Hubei, China | C. fioriniae | BK2-3 | Baokang, Hubei, China | |
| C. gloeosporioides | YY1-11 | Yunyang, Hubei, China | C. fioriniae | CX1 | Chengxian, Gansu, China | |
| C. fructicola | XS6 | Xingshan, Hubei, China | C. fioriniae | CX7 | Chengxian, Gansu, China | |
| C. fructicola | FX1-3 | Fengxiang, Shanxi, China | C. nymphaeae | ZS3-13 | Zhushan, Hubei, China | |
| C. fructicola | CX6 | Chengxian, Gansu, China | C. nymphaeae | ZS3-8 | Zhushan, Hubei, China | |
| C. fructicola | YY1-28 | Yunyang, Hubei, China | C. nymphaeae | BK1-11 | Baokang, Hubei, China | |
| C. fructicola | BK2-1 | Baokang, Hubei, China | C. nymphaeae | ZS2-6 | Zhushan, Hubei, China | |
| C. siamense | YX1-7 | Yunxi, Hubei, China | C. nymphaeae | XS5 | Xingshan, Hubei, China | |
| C. siamense | ZS2-9 | Zhushan, Hubei, China | C. nymphaeae | BK4-4 | Baokang, Hubei, China | |
| C. siamense | DJ3-5 | Danjiang, Hubei, China |
| Contig 编号 Contig code | 病毒名称 Name of putative viruses | 病毒名称缩写 Name abbreviation | 长度 Length | 序列相似度 Acid similarity/% | E值 E-value | 最佳匹配 Best match | 基因组类型 Genome type | 病毒科 Family | Contig 计数 Contig counts |
|---|---|---|---|---|---|---|---|---|---|
| Contig 22546 | Colletotrichum fioriniae alternavirus 1 | CfAV1 dsRNA2 | 2 463 bp | 70.4 | 3.50E-303 | Fusarium graminearum alternavirus 1(YP_009449446.1) | dsRNA | Alternaviridae | 1 |
| Contig11036 | Colletotrichum fioriniae alternavirus 1 | CfAV1 dsRNA1 | 3 534 bp | 73.3 | 0.00E+00 | Fusarium graminearum alternavirus 1(YP_009449439.1) | dsRNA | Alternaviridae | 1 |
| Contig22686 | Colletotrichum fioriniae alternavirus 1 | CfAV1 dsRNA3 | 2 454 bp | 72 | 3.00E-307 | Fusarium graminearum alternavirus 1(AUI80777.1) | dsRNA | Alternaviridae | 1 |
| Contig51809 | Colletotrichum fioriniae narnavirus 1 | CfNV1 | 1 136 nt | 59.28 | 3.00E-76 | Sclerotinia sclerotiorum narnavirus 2(QZE12028.1) | +ssRNA | Narnaviridae | 1 |
| Contig26097 | Colletotrichum siamense narnavirus 1 | CsNV1 | 2 239 nt | 65.3 | 0.00E+00 | Sclerotinia sclerotiorum narnavirus 2(QZE12027.1) | +ssRNA | Narnaviridae | 1 |
| Contig20122 | Colletotrichum siamense narnavirus 2 | CsNV2 | 2 420 nt | 33.2 | 8.00E-88 | Oidiodendron maius splipalmivirus 1(QNN89180.1) | +ssRNA | Narnaviridae | 1 |
| Contig11676 | Colletotrichum nymphaeae narnavirus 1 | CnNV1 | 3 090 nt | 65 | 0.00E+00 | Botryosphaeria dothidea narnavirus 3(QQD86177.1) | +ssRNA | Narnaviridae | 5 |
| Contig10410 | Colletotrichum fioriniae narnavirus 2 | CfNV2 | 3 619 nt | 55.3 | 0.00E+00 | Erysiphe necator associated narnavirus 15(QJT93747.1) | +ssRNA | Narnaviridae | 1 |
| Contig19559 | Colletotrichum nymphaeae narnavirus 2 | CnNV2 | 2 455 nt | 62.2 | 1.00E-278 | Erysiphe necator associated narnavirus 4(QHD64827.1) | +ssRNA | Narnaviridae | 2 |
| Contig4718 | Colletotrichum gloeosporioides narnavirus 1 | CgNV1 | 4 200 nt | 51.8 | 1.60E-300 | Monilinia narnavirus H (QED42934.1) | +ssRNA | Narnaviridae | 3 |
| Contig35776 | Colletotrichum fructicola narnavirus 1 | CfrNV1 | 1 689 nt | 60 | 1.00E-27 | Aspergillus lentulus narnavirus 1(BCH36644.1) | +ssRNA | Narnaviridae | 2 |
| Contig19474 | Colletotrichum fructicola botourmiavirus 1 | CfBV1 | 2 460 nt | 55.1 | 1.60E-178 | Pestalotiopsis botourmiavirus 3(QTH80197.1) | +ssRNA | Botourmiaviridae | 2 |
| Contig22684 | Colletotrichum fioriniae mitovirus 1 | CfMV1 | 2 455 nt | 51.7 | 4.00E-158 | Grapevine-associated mitovirus 10(QXN75363.1) | +ssRNA | Mitoviridae | 2 |
| Contig41113 | Colletotrichum fioriniae mitovirus 2 | CfMV2 | 1 493 nt | 78 | 9.10E-113 | Rhizoctonia solani mitovirus 33(QDW65423.1) | +ssRNA | Mitoviridae | 1 |
| Contig55502 | Colletotrichum fioriniae mitovirus 3 | CfMV3 | 1 012 nt | 68.2 | 1.50E-29 | Rhizoctonia solani mitovirus 33(QDW65423.1) | +ssRNA | Mitoviridae | 1 |
| Contig3026 | Colletotrichum gloeosporioides mitovirus 1 | CgMV1 | 4 742 nt | 91.4 | 4.20E-236 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 1 |
| Contig18460 | Colletotrichum fructicola mitovirus 1 | CfrMV1 | 2 520 nt | 94.5 | 0.00E+00 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 4 |
| Contig42866 | Colletotrichum siamense mitovirus 3 | CsMV3 | 1 462 nt | 93.1 | 4.20E-211 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig46248 | Colletotrichum fioriniae mitovirus 5 | CfMV4 | 1 337 nt | 89.4 | 2.90E-227 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig46871 | Colletotrichum gloeosporioides mitovirus 2 | CgMV2 | 1 313 nt | 92.4 | 1.30E-75 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig50030 | Colletotrichum nymphaeae mitovirus 1 | CnMV1 | 1 197 nt | 94.9 | 3.90E-66 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig23629 | Colletotrichum fioriniae mitovirus 6 | CfMV5 | 2 393 nt | 67.2 | 2.30E-259 | Colletotrichum falcatum mitovirus 1(AZT88621.1) | +ssRNA | Mitoviridae | 1 |
| Contig21451 | Colletotrichum siamense mitovirus 2 | CsMV2 | 2 539 nt | 65.2 | 1.20E-258 | Sclerotinia sclerotiorum mitovirus 32(AWY10990.1) | +ssRNA | Mitoviridae | 2 |
| Contig21504 | Colletotrichum siamense mitovirus 1 | CsMV1 | 2 536 nt | 65.5 | 1.5E-208 | Sclerotinia sclerotiorum mitovirus 6(QGY72611.1) | +ssRNA | Mitoviridae | 1 |
Table 2 Sequence information of sequenced viruses
| Contig 编号 Contig code | 病毒名称 Name of putative viruses | 病毒名称缩写 Name abbreviation | 长度 Length | 序列相似度 Acid similarity/% | E值 E-value | 最佳匹配 Best match | 基因组类型 Genome type | 病毒科 Family | Contig 计数 Contig counts |
|---|---|---|---|---|---|---|---|---|---|
| Contig 22546 | Colletotrichum fioriniae alternavirus 1 | CfAV1 dsRNA2 | 2 463 bp | 70.4 | 3.50E-303 | Fusarium graminearum alternavirus 1(YP_009449446.1) | dsRNA | Alternaviridae | 1 |
| Contig11036 | Colletotrichum fioriniae alternavirus 1 | CfAV1 dsRNA1 | 3 534 bp | 73.3 | 0.00E+00 | Fusarium graminearum alternavirus 1(YP_009449439.1) | dsRNA | Alternaviridae | 1 |
| Contig22686 | Colletotrichum fioriniae alternavirus 1 | CfAV1 dsRNA3 | 2 454 bp | 72 | 3.00E-307 | Fusarium graminearum alternavirus 1(AUI80777.1) | dsRNA | Alternaviridae | 1 |
| Contig51809 | Colletotrichum fioriniae narnavirus 1 | CfNV1 | 1 136 nt | 59.28 | 3.00E-76 | Sclerotinia sclerotiorum narnavirus 2(QZE12028.1) | +ssRNA | Narnaviridae | 1 |
| Contig26097 | Colletotrichum siamense narnavirus 1 | CsNV1 | 2 239 nt | 65.3 | 0.00E+00 | Sclerotinia sclerotiorum narnavirus 2(QZE12027.1) | +ssRNA | Narnaviridae | 1 |
| Contig20122 | Colletotrichum siamense narnavirus 2 | CsNV2 | 2 420 nt | 33.2 | 8.00E-88 | Oidiodendron maius splipalmivirus 1(QNN89180.1) | +ssRNA | Narnaviridae | 1 |
| Contig11676 | Colletotrichum nymphaeae narnavirus 1 | CnNV1 | 3 090 nt | 65 | 0.00E+00 | Botryosphaeria dothidea narnavirus 3(QQD86177.1) | +ssRNA | Narnaviridae | 5 |
| Contig10410 | Colletotrichum fioriniae narnavirus 2 | CfNV2 | 3 619 nt | 55.3 | 0.00E+00 | Erysiphe necator associated narnavirus 15(QJT93747.1) | +ssRNA | Narnaviridae | 1 |
| Contig19559 | Colletotrichum nymphaeae narnavirus 2 | CnNV2 | 2 455 nt | 62.2 | 1.00E-278 | Erysiphe necator associated narnavirus 4(QHD64827.1) | +ssRNA | Narnaviridae | 2 |
| Contig4718 | Colletotrichum gloeosporioides narnavirus 1 | CgNV1 | 4 200 nt | 51.8 | 1.60E-300 | Monilinia narnavirus H (QED42934.1) | +ssRNA | Narnaviridae | 3 |
| Contig35776 | Colletotrichum fructicola narnavirus 1 | CfrNV1 | 1 689 nt | 60 | 1.00E-27 | Aspergillus lentulus narnavirus 1(BCH36644.1) | +ssRNA | Narnaviridae | 2 |
| Contig19474 | Colletotrichum fructicola botourmiavirus 1 | CfBV1 | 2 460 nt | 55.1 | 1.60E-178 | Pestalotiopsis botourmiavirus 3(QTH80197.1) | +ssRNA | Botourmiaviridae | 2 |
| Contig22684 | Colletotrichum fioriniae mitovirus 1 | CfMV1 | 2 455 nt | 51.7 | 4.00E-158 | Grapevine-associated mitovirus 10(QXN75363.1) | +ssRNA | Mitoviridae | 2 |
| Contig41113 | Colletotrichum fioriniae mitovirus 2 | CfMV2 | 1 493 nt | 78 | 9.10E-113 | Rhizoctonia solani mitovirus 33(QDW65423.1) | +ssRNA | Mitoviridae | 1 |
| Contig55502 | Colletotrichum fioriniae mitovirus 3 | CfMV3 | 1 012 nt | 68.2 | 1.50E-29 | Rhizoctonia solani mitovirus 33(QDW65423.1) | +ssRNA | Mitoviridae | 1 |
| Contig3026 | Colletotrichum gloeosporioides mitovirus 1 | CgMV1 | 4 742 nt | 91.4 | 4.20E-236 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 1 |
| Contig18460 | Colletotrichum fructicola mitovirus 1 | CfrMV1 | 2 520 nt | 94.5 | 0.00E+00 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 4 |
| Contig42866 | Colletotrichum siamense mitovirus 3 | CsMV3 | 1 462 nt | 93.1 | 4.20E-211 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig46248 | Colletotrichum fioriniae mitovirus 5 | CfMV4 | 1 337 nt | 89.4 | 2.90E-227 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig46871 | Colletotrichum gloeosporioides mitovirus 2 | CgMV2 | 1 313 nt | 92.4 | 1.30E-75 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig50030 | Colletotrichum nymphaeae mitovirus 1 | CnMV1 | 1 197 nt | 94.9 | 3.90E-66 | Colletotrichum fructicola mitovirus 1(BBN51032.1) | +ssRNA | Mitoviridae | 5 |
| Contig23629 | Colletotrichum fioriniae mitovirus 6 | CfMV5 | 2 393 nt | 67.2 | 2.30E-259 | Colletotrichum falcatum mitovirus 1(AZT88621.1) | +ssRNA | Mitoviridae | 1 |
| Contig21451 | Colletotrichum siamense mitovirus 2 | CsMV2 | 2 539 nt | 65.2 | 1.20E-258 | Sclerotinia sclerotiorum mitovirus 32(AWY10990.1) | +ssRNA | Mitoviridae | 2 |
| Contig21504 | Colletotrichum siamense mitovirus 1 | CsMV1 | 2 536 nt | 65.5 | 1.5E-208 | Sclerotinia sclerotiorum mitovirus 6(QGY72611.1) | +ssRNA | Mitoviridae | 1 |
| 菌株Strain | 种名Species | 携带病毒名称Contained mycoviruses |
|---|---|---|
| BK1-11 | C. nymphaeae | CfMV4,CnNV2,CsNV1, CfAV1,CfMV1,CfMV2,CnMV1 |
| BK2-1 | C. fructicola | CfMV4,CsNV1,CfBV1,CfMV1,CfMV2 |
| BK2-3 | C. fioriniae | CfMV2,CfMV3,CfMV4,CfMV5,CfNV1,CfMV1 |
| BK3-5 | C. fioriniae | CfMV2,CfMV3,CfNV2,CfBV1 |
| BK4-4 | C. nymphaeae | CfMV2,CsMV3,CgMV2,CfAV1,CnNV1,CfMV1 |
| CX1 | C. fioriniae | CfMV2 |
| CX3 | C. fioriniae | CfMV2,CfMV4,CnNV1,CfMV1 |
| CX6 | C. fructicola | CfrMV1,CfMV4,CsMV2,CnNV1,CfBV1,CfMV1,CsNV1 |
| CX7 | C. fioriniae | CfAV1,CfMV1,CfMV3,CfMV2 |
| DJ1-1 | C. gloeosporioides | CfMV2,CfMV3 |
| DJ2-1 | C. gloeosporioide | CfrMV1,CgNV1,CsNV1,CnNV1,CfBV1,CfMV1,CfMV3,CfMV2 |
| DJ2-4 | C. gloeosporioide | CgMV1,CfrMV1,CfMV4,CfMV1,CfMV3,CfMV2,CfNV1 |
| DJ3-5 | C. siamense | CsMV3,CfMV4,CfMV5,CsMV2,CsNV1,CfNV1,CfMV1,CfMV3,CfMV2 |
| FX1-3 | C. fructicola | CsMV3,CfMV4,CfMV5,CsMV2,CfMV3,CfNV2,CsNV1,CfNV1,CfAV1,CfMV1,CfMV2 |
| SZ2-3 | C. gloeosporioides | CgMV2,CsMV2,CgNV1,CsNV1,CfNV1,CnNV1,CfMV1,CfMV2 |
| XS5 | C. nymphaeae | CsMV3,CfMV1,CfrMV1,CsMV1,CfrNV1,CfMV2 |
| XS6 | C. fructicola | CsMV3,CfNV2,CfrNV1,CsNV1,CnNV1,CfMV1,CfMV2 |
| YX1-5 | C. siamense | CfMV2,CgMV1,CfrMV1,CfMV4,CfMV5,CsMV2,CfNV1,CnNV1,CfBV1,CfMV1 |
| YX1-7 | C. siamense | CfMV2,CfrMV1,CfMV4,CfrNV1,CsNV2,CsNV1,CfAV1,CnNV1,CfMV1,CfBV1 |
| YY1-11 | C. gloeosporioides | CfMV3,CfMV2,CfrMV1,CfMV4,CgMV2,CfMV5,CsMV1,CnNV2,CsNV1,CnNV1,CfMV1 |
| YY1-28 | C. fructicola | CfrMV1,CfMV1,CfMV3,CfMV2 |
| ZS3-8 | C. nymphaeae | CfrNV1,CfMV1,CfMV2 |
| ZS2-6 | C. nymphaeae | CgMV2,CsMV2,CnNV2,CfrNV1,CsNV1,CfMV2 |
| ZS2-9 | C. siamense | CgMV1,CfMV4,CsMV2,CsMV1,CnNV2,CsNV2,CfAV1,CnNV1,CfBV1,CfMV1 |
| ZS3-13 | C. nymphaeae | CsNV1,CfMV2 |
Table 3 List of identification results of 22 mycoviruses in Colletotrichum spp. strains
| 菌株Strain | 种名Species | 携带病毒名称Contained mycoviruses |
|---|---|---|
| BK1-11 | C. nymphaeae | CfMV4,CnNV2,CsNV1, CfAV1,CfMV1,CfMV2,CnMV1 |
| BK2-1 | C. fructicola | CfMV4,CsNV1,CfBV1,CfMV1,CfMV2 |
| BK2-3 | C. fioriniae | CfMV2,CfMV3,CfMV4,CfMV5,CfNV1,CfMV1 |
| BK3-5 | C. fioriniae | CfMV2,CfMV3,CfNV2,CfBV1 |
| BK4-4 | C. nymphaeae | CfMV2,CsMV3,CgMV2,CfAV1,CnNV1,CfMV1 |
| CX1 | C. fioriniae | CfMV2 |
| CX3 | C. fioriniae | CfMV2,CfMV4,CnNV1,CfMV1 |
| CX6 | C. fructicola | CfrMV1,CfMV4,CsMV2,CnNV1,CfBV1,CfMV1,CsNV1 |
| CX7 | C. fioriniae | CfAV1,CfMV1,CfMV3,CfMV2 |
| DJ1-1 | C. gloeosporioides | CfMV2,CfMV3 |
| DJ2-1 | C. gloeosporioide | CfrMV1,CgNV1,CsNV1,CnNV1,CfBV1,CfMV1,CfMV3,CfMV2 |
| DJ2-4 | C. gloeosporioide | CgMV1,CfrMV1,CfMV4,CfMV1,CfMV3,CfMV2,CfNV1 |
| DJ3-5 | C. siamense | CsMV3,CfMV4,CfMV5,CsMV2,CsNV1,CfNV1,CfMV1,CfMV3,CfMV2 |
| FX1-3 | C. fructicola | CsMV3,CfMV4,CfMV5,CsMV2,CfMV3,CfNV2,CsNV1,CfNV1,CfAV1,CfMV1,CfMV2 |
| SZ2-3 | C. gloeosporioides | CgMV2,CsMV2,CgNV1,CsNV1,CfNV1,CnNV1,CfMV1,CfMV2 |
| XS5 | C. nymphaeae | CsMV3,CfMV1,CfrMV1,CsMV1,CfrNV1,CfMV2 |
| XS6 | C. fructicola | CsMV3,CfNV2,CfrNV1,CsNV1,CnNV1,CfMV1,CfMV2 |
| YX1-5 | C. siamense | CfMV2,CgMV1,CfrMV1,CfMV4,CfMV5,CsMV2,CfNV1,CnNV1,CfBV1,CfMV1 |
| YX1-7 | C. siamense | CfMV2,CfrMV1,CfMV4,CfrNV1,CsNV2,CsNV1,CfAV1,CnNV1,CfMV1,CfBV1 |
| YY1-11 | C. gloeosporioides | CfMV3,CfMV2,CfrMV1,CfMV4,CgMV2,CfMV5,CsMV1,CnNV2,CsNV1,CnNV1,CfMV1 |
| YY1-28 | C. fructicola | CfrMV1,CfMV1,CfMV3,CfMV2 |
| ZS3-8 | C. nymphaeae | CfrNV1,CfMV1,CfMV2 |
| ZS2-6 | C. nymphaeae | CgMV2,CsMV2,CnNV2,CfrNV1,CsNV1,CfMV2 |
| ZS2-9 | C. siamense | CgMV1,CfMV4,CsMV2,CsMV1,CnNV2,CsNV2,CfAV1,CnNV1,CfBV1,CfMV1 |
| ZS3-13 | C. nymphaeae | CsNV1,CfMV2 |
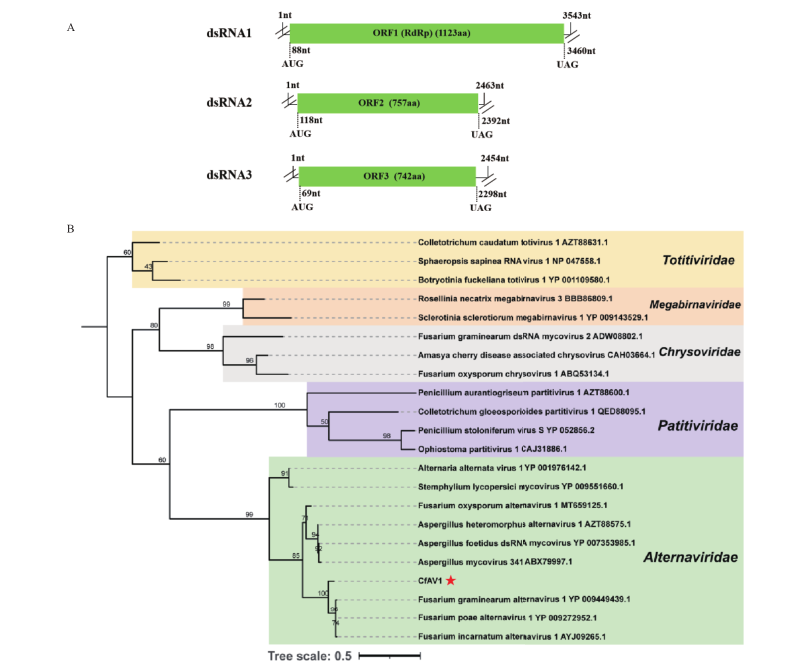
Fig. 2 Genome organization of 3 dsRNA segments in CfAV1and phylogenetic analysis of viruses in family Alternaviridae and other related viruses A: Genome organization of 3 dsRNA segments in CfAV1. B: A maximum likelihood phylogenetic tree was constructed based on the RdRp sequence of CfAV1 and other related viruses. CfAV1is marked with red star
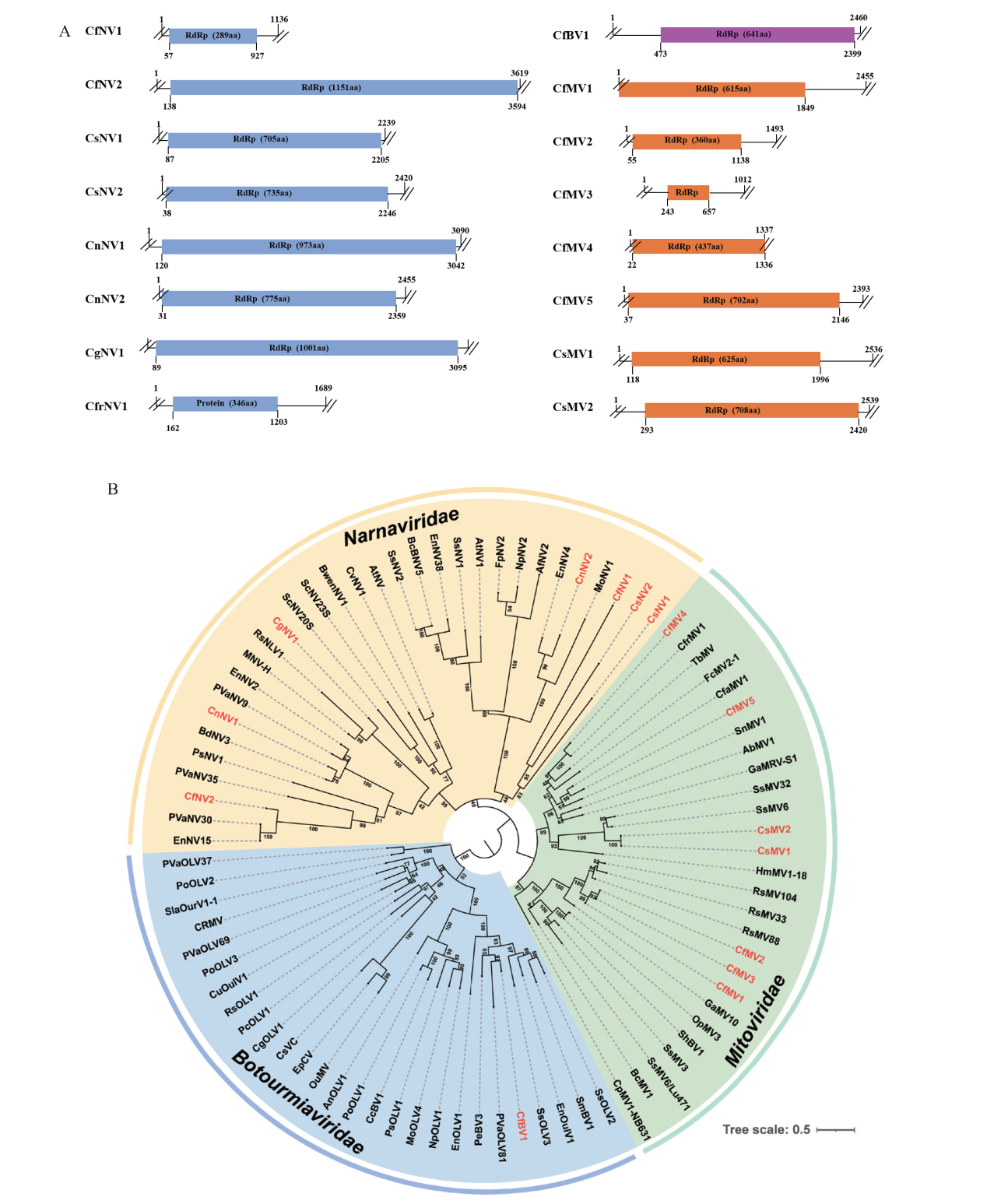
Fig. 3 Genome organization and phylogenetic analysis of viruses in family Narnaviridae, Mitoviridae, and Botourmiaviridae A: Genome organization of novel viruses in family Narnaviridae, Mitoviridae, and Botourmiaviridae. B: A maximum likelihood phylogenetic tree constructed based on the amino acid sequences of viral RdRp. The numbers next to each branch refers to the bootstrap support based on 1 000 replicates. Viruses identified in this study are denoted by the abbreviated names
| [1] |
Ghabrial SA, Suzuki N. Viruses of plant pathogenic fungi[J]. Annu Rev Phytopathol, 2009, 47: 353-384.
doi: 10.1146/annurev-phyto-080508-081932 pmid: 19400634 |
| [2] |
Xie JT, Jiang DH. New insights into mycoviruses and exploration for the biological control of crop fungal diseases[J]. Annu Rev Phytopathol, 2014, 52: 45-68.
doi: 10.1146/annurev-phyto-102313-050222 pmid: 25001452 |
| [3] | Dolja VV, Koonin EV. Capsid-less RNA viruses[M]. In:eLS ed.ed. Chichester: John Wiley & Sons, 2012. |
| [4] |
Son M, Yu J, Kim KH. Five questions about mycoviruses[J]. PLoS Pathog, 2015, 11(11): e1005172.
doi: 10.1371/journal.ppat.1005172 URL |
| [5] |
Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface[J]. Nat Rev Microbiol, 2005, 3(8): 632-642.
pmid: 16064055 |
| [6] |
Anagnostakis SL. Biological control of chestnut blight[J]. Science, 1982, 215(4532): 466-471.
pmid: 17771259 |
| [7] |
Yu X, Li B, Fu Y, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus[J]. PNAS, 2010, 107(18): 8387-8392.
doi: 10.1073/pnas.0913535107 pmid: 20404139 |
| [8] |
Yu X, Li B, Fu Y, et al. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide[J]. PNAS, 2013, 110(4): 1452-1457.
doi: 10.1073/pnas.1213755110 pmid: 23297222 |
| [9] |
Dean R, Kan JAV, Pretorius ZA, et al. The Top 10 fungal pathogens in molecular plant pathology[J]. Mol Plant Pathol, 2012, 13(4): 414-430.
doi: 10.1111/j.1364-3703.2011.00783.x pmid: 22471698 |
| [10] |
Zhong J, Chen D, Lei XH, et al. Detection and characterization of a novel Gammapartitivirus in the phytopathogenic fungus Colletotrichum acutatum strain HNZJ001[J]. Virus Res, 2014, 190: 104-109.
doi: 10.1016/j.virusres.2014.05.028 pmid: 25008759 |
| [11] |
Wang Y, Liu S, Zhu HJ, et al. Molecular characterization of a novel mycovirus from the plant pathogenic fungus Colletotrichum gloeosporioides[J]. Arch Virol, 2019, 164(11): 2859-2863.
doi: 10.1007/s00705-019-04354-2 |
| [12] |
Marzano SYL, Nelson BD, Ajayi-Oyetunde O, et al. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens[J]. J Virol, 2016, 90(15): 6846-6863.
doi: 10.1128/JVI.00357-16 URL |
| [13] |
Campo S, Gilbert KB, Carrington JC. Small RNA-based antiviral defense in the phytopathogenic fungus Colletotrichum higginsianum[J]. PLoS Pathog, 2016, 12(6): e1005640.
doi: 10.1371/journal.ppat.1005640 URL |
| [14] |
Zhong J, Pang XD, Zhu HJ, et al. Molecular characterization of a trisegmented mycovirus from the plant pathogenic fungus Colletotrichum gloeosporioides[J]. Viruses, 2016, 8(10): 268.
doi: 10.3390/v8100268 URL |
| [15] |
Zhai LF, Zhang MX, Hong N, et al. Identification and characterization of a novel hepta-segmented dsRNA virus from the phytopathogenic fungus Colletotrichum fructicola[J]. Front Microbiol, 2018, 9: 754.
doi: 10.3389/fmicb.2018.00754 URL |
| [16] |
Jia HX, Dong KL, Zhou LL, et al. A dsRNA virus with filamentous viral particles[J]. Nat Commun, 2017, 8(1): 168.
doi: 10.1038/s41467-017-00237-9 pmid: 28761042 |
| [17] |
Guo J, Zhu JZ, Zhou XY, et al. A novel ourmia-like mycovirus isolated from the plant pathogenic fungus Colletotrichum gloeosporioi-des[J]. Arch Virol, 2019, 164(10): 2631-2635.
doi: 10.1007/s00705-019-04346-2 |
| [18] |
Xu XW, Hai D, Li JC, et al. Molecular characterization of a novel penoulivirus from the phytopathogenic fungus Colletotrichum camelliae[J]. Arch Virol, 2022, 167(2): 641-644.
doi: 10.1007/s00705-021-05334-1 |
| [19] |
Liu H, Wang H, Lu X, et al. Molecular characterization of a novel single-stranded RNA virus, ChRV1, isolated from the plant pathogenic fungus Colletotrichum Higginsianum[J]. Archives of Virology, 2021, 166(6):1805-1809.
doi: 10.1007/s00705-021-05071-5 |
| [20] |
Wang H, Liu H, Zhou Q. The complete genome sequence of a new Mitovirus from the phytopathogenic fungus Colletotrichum higginsianum[J]. Arch Virol, 2021, 166(5): 1481-1484.
doi: 10.1007/s00705-021-04996-1 pmid: 33616726 |
| [21] |
Gilbert KB, Holcomb EE, Allscheid RL, et al. Hiding in plain sight: new virus genomes discovered via a systematic analysis of fungal public transcriptomes[J]. PLoS One, 2019, 14(7): e0219207.
doi: 10.1371/journal.pone.0219207 URL |
| [22] | Lima JS, Figueiredo JG, Gomes RG, et al. Genetic diversity of Colletotrichum spp. an endophytic fungi in a medicinal plant, Brazilian pepper tree[J]. ISRN Microbiol, 2012, 2012: 215716. |
| [23] |
Figueiredo LC, Figueiredo GS, Giancoli AC, et al. Detection of isometric, dsRNA-containing viral particles in Colletotrichum gloeosporioides isolated from cashew tree[J]. Tropical Plant Pathology, 2012, 37(2): 142-145.
doi: 10.1590/S1982-56762012000200007 URL |
| [24] |
Zhang HX, Xie JT, Fu YP, et al. A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for Brassica protection and yield enhancement[J]. Mol Plant, 2020, 13(10): 1420-1433.
doi: 10.1016/j.molp.2020.08.016 URL |
| [25] |
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120.
doi: 10.1093/bioinformatics/btu170 pmid: 24695404 |
| [26] |
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements[J]. Nat Methods, 2015, 12(4): 357-360.
doi: 10.1038/nmeth.3317 pmid: 25751142 |
| [27] |
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079.
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [28] |
Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing[J]. J Comput Biol, 2012, 19(5): 455-477.
doi: 10.1089/cmb.2012.0021 pmid: 22506599 |
| [29] |
Fu LM, Niu BF, Zhu ZW, et al. CD-HIT: accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152.
doi: 10.1093/bioinformatics/bts565 pmid: 23060610 |
| [30] |
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND[J]. Nat Methods, 2015, 12(1): 59-60.
doi: 10.1038/nmeth.3176 pmid: 25402007 |
| [31] |
Nakamura T, Yamada KD, Tomii K, et al. Parallelization of MAFFT for large-scale multiple sequence alignments[J]. Bioinformatics, 2018, 34(14): 2490-2492.
doi: 10.1093/bioinformatics/bty121 pmid: 29506019 |
| [32] |
Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates[J]. Nat Methods, 2017, 14(6): 587-589.
doi: 10.1038/nmeth.4285 pmid: 28481363 |
| [33] |
Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies[J]. Mol Biol Evol, 2015, 32(1): 268-274.
doi: 10.1093/molbev/msu300 URL |
| [34] |
Sutela S, Forgia M, Vainio EJ, et al. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization[J]. Virus Evol, 2020, 6(2): veaa076.
doi: 10.1093/ve/veaa076 URL |
| [35] |
Poimala A, Parikka P, Hantula J, et al. Viral diversity in Phytophthora cactorum population infecting strawberry[J]. Environ Microbiol, 2021, 23(9): 5200-5221.
doi: 10.1111/1462-2920.15519 pmid: 33848054 |
| [36] |
Wen CY, Wan XR, Zhang YY, et al. Molecular characterization of the first alternavirus identified in Fusarium oxysporum[J]. Viruses, 2021, 13(10): 2026.
doi: 10.3390/v13102026 URL |
| [37] |
Aoki N, Moriyama H, Kodama M, et al. A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata[J]. Virus Res, 2009, 140(1-2): 179-187.
doi: 10.1016/j.virusres.2008.12.003 URL |
| [38] |
Hillman BI, Cai GH. The family Narnaviridae: simplest of RNA viruses[J]. Adv Virus Res, 2013, 86: 149-176.
doi: 10.1016/B978-0-12-394315-6.00006-4 pmid: 23498906 |
| [39] | Chiba S. Fusariviruses[M]. In: Bamford DH and Zuckerman M eds. Encyclopedia of virology. Oxford: Academic Press, 2021: 577-581. |
| [40] | Ruiz-Padilla A, Rodríguez-Romero J, Gómez-Cid I, et al. Novel mycoviruses discovered in the mycovirome of a necrotrophic fungus[J]. mBio, 2021, 12(3): e03705-e03720. |
| [41] |
Turina M, Hillman BI, Izadpanah K, et al. ICTV virus taxonomy profile: Ourmiavirus[J]. The Journal of General Virology, 2017, 98(2):129-130.
doi: 10.1099/jgv.0.000725 URL |
| [42] |
Ayllón MA, Turina M, Xie J, et al. ICTV virus taxonomy profile: Botourmiaviridae[J]. J Gen Virol, 2020, 101(5): 454-455.
doi: 10.1099/jgv.0.001409 pmid: 32375992 |
| [1] | YU Yang, LIU Tian-hai, LIU Li-xu, TANG Jie, PENG Wei-hong, CHEN Yang, TAN Hao. Study on Aerosol Microbial Community in the Production Workshop of Morel Spawn [J]. Biotechnology Bulletin, 2023, 39(5): 267-275. |
| [2] | LI Shan-jia, LEI Yu-xin, SUN Meng-ge, LIU Hai-feng, WANG Xing-min. Research Progress in the Diversity of Endophytic Bacteria in Seeds and Their Interaction with Plants [J]. Biotechnology Bulletin, 2023, 39(4): 166-175. |
| [3] | GUO Wen-bo, LU Yang, SUI Li, ZHAO Yu, ZOU Xiao-wei, ZHANG Zheng-kun, LI Qi-yun. Preparation and Application of Polyclonal Antibodies Against Beauveria bassiana Mycovirus BbPmV-4 Coat Protein [J]. Biotechnology Bulletin, 2023, 39(10): 58-67. |
| [4] | WANG Zi-ye, WANG Zhi-gang, YAN Ai-hua. Diversity of Soil Protist Community in the Rhizosphere of Morus alba L. at Different Tree Ages [J]. Biotechnology Bulletin, 2022, 38(8): 206-215. |
| [5] | GAO Xiao-ning, LIU Rui, WU Zi-lin, WU Jia-yun. Characteristics of Endophytic Fungal and Bacterial Community in the Stalks of Sugarcane Cultivars Resistant to Ratoon Stunting Disease [J]. Biotechnology Bulletin, 2022, 38(6): 166-173. |
| [6] | XU Yang, ZHANG Guan-chu, DING Hong, QIN Fei-fei, ZHANG Zhi-meng, DAI Liang-xiang. Effects of Soil Types on Bacterial Community Diversity on the Rhizosphere Soil of Arachis hypogaea and Yield [J]. Biotechnology Bulletin, 2022, 38(6): 221-234. |
| [7] | ZHONG Hui, LIU Ya-jun, WANG Bin-hua, HE Meng-jie, WU Lan. Effects of Analysis Methods on the Analyzed Results of 16S rRNA Gene Amplicon Sequencing in Bacterial Communities [J]. Biotechnology Bulletin, 2022, 38(6): 81-92. |
| [8] | ZHOU Xiao-nan, XU Jin-qing, LEI Yu-qing, WANG Hai-qing. Development of SNP Markers in Medicago archiducis-nicolai Based on GBS-seq [J]. Biotechnology Bulletin, 2022, 38(4): 303-310. |
| [9] | XIE Guo-zhen, TANG Yuan, NING Xiao-mei, QIU Ji-hui, TAN Zhou-jin. Effects of Dendrobium officinale Polysaccharides on the Intestinal Mucosal Structure and Microbiota in Mice Fed a High-fat Diet [J]. Biotechnology Bulletin, 2022, 38(2): 150-157. |
| [10] | LIU Shuang, YAO Jia-ni, SHEN Cong, DAI Jin-xia. Fluorescent Quantitative PCR of nifH Gene and Diversity Analysis of Nitrogen-fixing Bacteria in the Rhizosphere Soil of Caragana spp. of Desert Grassland [J]. Biotechnology Bulletin, 2022, 38(12): 252-262. |
| [11] | CHEN Yu-jie, ZHENG Hua-bao, ZHOU Xin-yan. Modified High-throughput Sequencing Reveals the Effects of Different Algicides towards Algal Community [J]. Biotechnology Bulletin, 2022, 38(11): 70-79. |
| [12] | LI Ting-ting, DENG Xu-hui, LI Ruo-chen, LIU Hong-jun, SHEN Zong-zhuan, LI Rong, SHEN Qi-rong. Effects of Ralstonia solanacearum Infection on Soil Fungal Community Diversity [J]. Biotechnology Bulletin, 2022, 38(10): 195-203. |
| [13] | YAN Hui-lin, LU Guang-xin, DENG Ye, GU Song-song, YAN Cheng-liang, MA Kun, ZHAO Yang-an, ZHANG Hai-juan, WANG Ying-cheng, ZHOU Xue-li, DOU Sheng-yun. Effects of Rhizobium Seed Dressing on the Soil Microbial Community of Grass-legume Mixtures in Alpine Regions [J]. Biotechnology Bulletin, 2022, 38(10): 204-215. |
| [14] | ZHANG Tian-tian, LI Yong-zhen, SHEN Guo-ping, WANG Rong, ZHU De-rui, XING Jiang-wa. Population Diversity of Isolated Halophilic and Halotolerant Bacteria from Hypersaline Salt Lakes and Evaluation of Ectoine Production [J]. Biotechnology Bulletin, 2022, 38(1): 168-178. |
| [15] | WANG Zhi-shan, LI Ni, WANG Wei-ping, LIU Yang. Research Progress in Endophytic Bacteria in Rice Seeds [J]. Biotechnology Bulletin, 2022, 38(1): 236-246. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||