








生物技术通报 ›› 2021, Vol. 37 ›› Issue (11): 212-224.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0108
收稿日期:2021-01-27
出版日期:2021-11-26
发布日期:2021-12-03
作者简介:张云川,男,硕士研究生,研究方向:生物化学与分子生物学;E-mail: 基金资助:
ZHANG Yun-chuan( ), LIN Yi-xuan, CAO Xin-wen, WANG Hai-nan, YAN Jie(
), LIN Yi-xuan, CAO Xin-wen, WANG Hai-nan, YAN Jie( )
)
Received:2021-01-27
Published:2021-11-26
Online:2021-12-03
摘要:
为探究茉莉酸在提高植物抗旱性方面发挥的重要作用。对橡胶草外施茉莉酸甲酯(MeJA)处理的转录组进行差异表达谱分析,并筛选克隆出一个受茉莉酸诱导表达显著上调的AP2/EREBP亚家族基因DREB,命名为TkDREB2,对其进行生物信息学分析并转化烟草,进一步对TkDREB2转基因烟草植株在发育期的抗旱性功能鉴定,对转基因烟草进行自然干旱处理,并测定相关生理指标。结果显示在茉莉酸甲酯处理的转录组分析中发现与茉莉酸合成相关的基因和转录因子AP2/EREBP、MYB、MYC、NAC和WRKY的表达水平发生显著变化。筛选克隆的TkDREB2基因全长CDS序列为1 002 bp,编码333个氨基酸,属于AP2/EREBP亚家族的DREB型转录因子;系统进化树分析表明,该蛋白与苜蓿的DREB蛋白同源性最近。对转基因烟草进行自然干旱处理15 d后发现野生型叶片开始萎蔫;继续处理至22 d,野生型叶片严重萎蔫而转基因烟草仅轻微萎蔫,测定正常生长及干旱处理后野生型和转基因烟草的相关生理指标发现:在正常生长条件下,转基因烟草的超氧化物歧化酶(SOD)、过氧化物酶(POD)和过氧化氢酶(CAT)活性大约是野生型的2倍;在干旱处理后,转基因烟草的CAT、SOD和POD活性分别是野生型的2倍、2倍和1.5倍左右,丙二醛(MDA)含量和相对电导率仅是野生型的1/2左右。综合以上结果表明TkDREB2能够积极响应茉莉酸的诱导表达,过表达TkDREB2基因能够明显提高烟草的抗旱性,推测DREB2转录因子可能在茉莉酸信号转导调控植物抗旱性方面发挥重要作用。
张云川, 林熠轩, 曹新文, 王海楠, 闫洁. 橡胶草TkDREB2基因的克隆以及在烟草中的抗旱功能分析[J]. 生物技术通报, 2021, 37(11): 212-224.
ZHANG Yun-chuan, LIN Yi-xuan, CAO Xin-wen, WANG Hai-nan, YAN Jie. TkDREB2 Clone from Taraxacum kok-saghyz and Drought Tolerance Analysis of Transgenic Nicotiana tabacum[J]. Biotechnology Bulletin, 2021, 37(11): 212-224.

图1 差异表达的AP2/EREBP(A)、bHLH(B)、MYB(C)和WRKY(D)转录因子热图 红色表达上调,蓝色表达下调,颜色越深代表表达量越高;MJ0_R、MJ6_R和MJ24_R表示茉莉酸处理0、6和24 h
Fig. 1 Heat map of differentially expressed AP2/EREBP(A),bHLH(B),MYB(C)and WRKY(D)transcription factors Red expression is up-regulated,blue expression is down-regulated,the darker the color,the higher the expression level. MJ0_R,MJ6_R and MJ24_R indicate jasmonic acid treatment 0,6 and 24 h
| 基因信息Genetic information | 茉莉酸处理时间Treatment time by jasmonic acid | 差异表达倍数Log2FoldChange | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 基因名称 Gene name | 基因Gene ID | MJ6 | MJ0 | MJ24 | 6 VS 0 | P value | 24 VS 0 | P value | |||
| OPR | c49640_g1 | 25 210.72 | 956.65 | 4.72 | 1.71E-35 | ||||||
| 1 017.64 | 16 350.86 | 4.01 | 1.77E-31 | ||||||||
| c49640_g2 | 15 311.67 | 589.34 | 4.70 | 3.61E-35 | |||||||
| 626.95 | 10 019.59 | 4.00 | 3.15E-33 | ||||||||
| c52666_g1 | 2 642.99 | 268.00 | 3.30 | 1.82E-19 | |||||||
| 284.96 | 2 252.14 | 2.98 | 4.39E-20 | ||||||||
| AOS | c39329_g1 | 1 825.74 | 17.73 | 6.69 | 2.28E-17 | ||||||
| 18.87 | 10 973.08 | 9.18 | 3.60E-25 | ||||||||
| c47723_g1 | 603.38 | 7 806.60 | 3.69 | 1.46E-06 | |||||||
| LOX | c53508_g1 | 21 905.05 | 359.65 | 5.93 | 4.42E-10 | ||||||
| 382.62 | 49 808.79 | 7.02 | 2.75E-13 | ||||||||
| c51038_g1 | 294.97 | 895.27 | 1.60 | 1.62E-05 | |||||||
| AOC | c44998_g1 | 369.99 | 146.00 | 1.34 | 0.000 269 | ||||||
| 155.26 | 737.86 | 2.25 | 9.16E-12 | ||||||||
表1 茉莉酸合成相关酶编码基因的差异表达情况
Table 1 Differential expression of genes encoding enzymes related to jasmonic acid synthesis
| 基因信息Genetic information | 茉莉酸处理时间Treatment time by jasmonic acid | 差异表达倍数Log2FoldChange | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 基因名称 Gene name | 基因Gene ID | MJ6 | MJ0 | MJ24 | 6 VS 0 | P value | 24 VS 0 | P value | |||
| OPR | c49640_g1 | 25 210.72 | 956.65 | 4.72 | 1.71E-35 | ||||||
| 1 017.64 | 16 350.86 | 4.01 | 1.77E-31 | ||||||||
| c49640_g2 | 15 311.67 | 589.34 | 4.70 | 3.61E-35 | |||||||
| 626.95 | 10 019.59 | 4.00 | 3.15E-33 | ||||||||
| c52666_g1 | 2 642.99 | 268.00 | 3.30 | 1.82E-19 | |||||||
| 284.96 | 2 252.14 | 2.98 | 4.39E-20 | ||||||||
| AOS | c39329_g1 | 1 825.74 | 17.73 | 6.69 | 2.28E-17 | ||||||
| 18.87 | 10 973.08 | 9.18 | 3.60E-25 | ||||||||
| c47723_g1 | 603.38 | 7 806.60 | 3.69 | 1.46E-06 | |||||||
| LOX | c53508_g1 | 21 905.05 | 359.65 | 5.93 | 4.42E-10 | ||||||
| 382.62 | 49 808.79 | 7.02 | 2.75E-13 | ||||||||
| c51038_g1 | 294.97 | 895.27 | 1.60 | 1.62E-05 | |||||||
| AOC | c44998_g1 | 369.99 | 146.00 | 1.34 | 0.000 269 | ||||||
| 155.26 | 737.86 | 2.25 | 9.16E-12 | ||||||||

图3 重组质粒pMD19T-TkDREB2酶切 M:DNA marker Ⅲ;1-2:BamHI和Pst I双酶切pMD19T-TkDREB2重组质粒;3:pMD19T-TkDREB2重组质粒
Fig. 3 Recombinant plasmid pMD19T-TkDREB2 digestion M: DNA marker Ⅲ. 1-2: BamHI and Pst I double digestion pMD19T-TkDREB2 recombinant plasmid. 3: pMD19T-TkDREB2 recombinant plasmid

图6 TkDREB2蛋白二级结构预测 蓝色:α-螺旋;红色:延伸链;绿色:β-转角;紫色:无规则卷曲
Fig. 6 Prediction of the secondary structure of TkDREB2 protein Blue:α-helix;red:extended chain;green:β-turn;purple:random curl
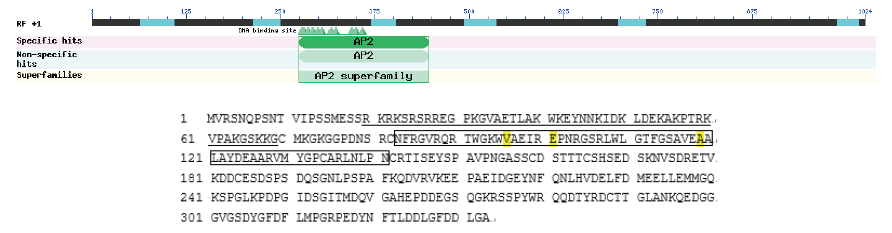
图8 TkDREB2氨基酸序列分析 方框内的序列代表AP2结构域;下划线部分表示预测的核定位信号序列
Fig. 8 TkDREB2 amino acid sequence analysis The sequence in the square represents the AP2 domain, and the underlined part represents the predicted nuclear localization signal sequence

图10 TkDREB2蛋白系统进化树和保守motif分析 A:TkDREB2蛋白系统进化树和保守motif分析;B:5个保守motif片段。Taraxacum kok-saghyz:TkDREB2(橡胶草,ASL72071.1);Lactuca sativa:LsDREB2A(莴苣,XP_023748594.1);Cichorium intybus:CiDREB2(菊苣,AHJ08574.1);Cynara cardunculus:CcDREB2A(朝鲜蓟,XP_024968974.1);Chrysanthemum vestitum:CvDREB2(毛华菊,ABR23508.1);Helianthus annuus:HaDREB2A(向日葵,XP_021982098.1);Nicotiana sylvestris:NsDREB2A(美花烟草,XP_009788693.1);Nicotiana attenuata:NaDREB2A(狼烟草,XP_019245341.1);Nicotiana tomentosiformis:NtDREB2A(茸毛烟草,XP_009594350.1);Nicotiana tabacum:NtDREB2A(普通烟草,XP_016461559.1);Sesamum indicum:SiDREB2C(芝麻,XP_011084866.1)
Fig. 10 Phylogenetic tree and conservative motif analysis of TkDREB2 protein A:Phylogenetic tree and conservative motif analysis of TkDREB2 protein. B:5 conservative motif fragments. Taraxacum kok-saghyz:TkDREB2(Taraxacum kok-saghyz,ASL72071.1); Lactuca sativa:LsDREB2A(Lactuca sativa,XP_023748594.1); Cichorium intybus:CiDREB2(Cichorium intybus,AHJ08574.1); Cynara cardunculus:CcDREB2A(Cynara cardunculus,XP_024968974.1); Chrysanthemum vestitum:CvDREB2(Chrysanthemum vestitum,ABR23508.1); Helianthus annuus:HaDREB2A(Helianthus annuus,XP_021982098.1); Nicotiana sylvestris:NsDREB2A(Nicotiana sylvestris,XP_009788693.1); Nicotiana attenuata:NaDREB2A(Nicotiana attenuata,XP_019245341.1); Nicotiana tomentosiformis:NtDREB2A(Nicotiana tomentosiformis,XP_009594350.1); Nicotiana tabacum:NtDREB2A(Nicotiana tabacum,XP_016461559.1); Sesamum indicum:SiDREB2C(Sesamum indicum,XP_011084866.1)

图11 重组质粒pCAMBIA2300-35S-TkDREB2酶切 M:DNA marker Ⅲ;1:pCAMBIA2300-35S-TkDREB2重组质粒;2-3:BamHI和Pst I双酶切pCAMBIA2300-35S-TkDREB2重组质粒
Fig. 11 Recombinant plasmid pCAMBIA2300-35S-TkDREB2 digestion M: DNA marker Ⅲ; 1: pCAMBIA2300-35S-TkDREB2 recombinant plasmid;2-3: BamHI and Pst I double digestion pCAMBIA2300-35S-TkDREB2 recombinant plasmid

图12 PCR鉴定TkDREB2转基因植株 M:DNA marker Ⅲ;2-9:8个鉴定正确的转基因植株
Fig. 12 PCR identification of TkDREB2 transgenic plants M: DNA marker Ⅲ; 2-9: 8 correctly identified transgenic plants

图13 正常生长条件(A)和干旱处理22 d(B)的野生型(WT)与转基因烟草 Transgenic lines:转基因烟草;WT:野生型烟草
Fig. 13 Wild-type(WT)and genetically modified tobacco under normal growth conditions(A)and 22 d of drought treatment(B) Transgenic lines:Genetically modified tobacco;WT:wild type tobacco
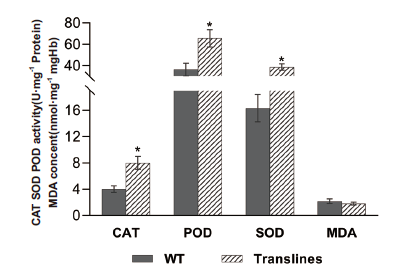
图14 正常生长条件下野生型和转基因植株的SOD、POD和CAT酶活以及MDA含量测定结果 3株野生型和3株转基因烟草各生理指标测定结果的均值作图进行比较;*表示与野生型相比差异显著(P<0.05,t-test),下同
Fig. 14 SOD,POD and CAT enzyme activities and MDA content measurement results of wild-type and transgenic plants under normal growth conditions The mean measurement results of the physiological indexes of 3 wild-type and 3 transgenic tobaccos are compared by plotting. * indicates significant difference compared with wild-type(P<0.05,t-test),the same below

图15 干旱处理后野生型和转基因植株的SOD、POD和CAT酶活以及MDA含量测定结果
Fig. 15 SOD,POD and CAT enzyme activities and MDA content measurement results of wild-type and transgenic plants after drought treatment

图16 正常生长和干旱处理后野生型和转基因植株电导率测定结果 Transline 1-3:野生型植株的电导率值是3株野生型测定结果的均值
Fig. 16 Electrical conductivity measurement results of wild-type and transgenic plants after normal growth and drought treatment Transline 1-3:3 strains of genetically modified tobacco. The electrical conductivity of the wild-type plant is the mean value of the measurement results of 3 wild-type plants
| [1] |
Chini A, Fonseca S, Fernández G, et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature, 2007, 448(7154): 666-671.
doi: 10.1038/nature06006 URL |
| [2] |
Fonseca S, Chini A, Hamberg M, et al. (+)-7-Iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate[J]. Nat Chem Biol, 2009, 5(5): 344-350.
doi: 10.1038/nchembio.161 pmid: 19349968 |
| [3] |
Westfall CS, Zubieta C, Herrmann J, et al. Structural basis for prereceptor modulation of plant hormones by GH3 proteins[J]. Science, 2012, 336(6089): 1708-1711.
doi: 10.1126/science.1221863 pmid: 22628555 |
| [4] |
Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling[J]. Nature, 2010, 464(7289): 788-791.
doi: 10.1038/nature08854 URL |
| [5] | 文锦芬, 赵凯, 邓明华. 转录因子DREB、ERF和NAC在介导植物响应生物和非生物胁迫中的作用[J]. 湖南生态科学学报, 2019, 6(3): 51-59. |
| Wen JF, Zhao K, Deng MH. Roles of transcription factors DREB, ERF and NAC in mediating plant responses to biotic and abiotic stresses[J]. J Hunan Ecol Sci, 2019, 6(3): 51-59. | |
| [6] | 王舟, 刘建秀. DREB/CBF类转录因子研究进展及其在草坪草和牧草抗逆基因工程中的应用[J]. 草业学报, 2011, 20(1): 222-236. |
| Wang Z, Liu JX. Advances in studies on DREB/CBF transcription factors, and their applications in genetic engineering for stress tolerance of turf and forage grasses[J]. Acta Prataculturae Sin, 2011, 20(1): 222-236. | |
| [7] |
Hussain SS, Kayani MA, Amjad M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants[J]. Biotechnol Prog, 2011, 27(2): 297-306.
doi: 10.1002/btpr.v27.2 URL |
| [8] |
Zhang S, Zhu C, Lyu Y, et al. Genome-wide identification, molecular evolution, and expression analysis provide new insights into the APETALA2/ethylene responsive factor(AP2/ERF)superfamily in Dimocarpus longan Lour[J]. BMC Genomics, 2020, 21(1): 62.
doi: 10.1186/s12864-020-6469-4 URL |
| [9] |
Aya K, Hobo T, et al. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway[J]. Plant Cell Physiol, 2014, 55(5): 897-912.
doi: 10.1093/pcp/pcu023 URL |
| [10] |
Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana[J]. Science, 2003, 301(5633): 653-657.
pmid: 12893945 |
| [11] |
Hu YX, Wang YH, Liu XF, et al. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development[J]. Cell Res, 2004, 14(1): 8-15.
doi: 10.1038/sj.cr.7290197 URL |
| [12] |
Sakuma Y, Liu Q, Dubouzet JG, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression[J]. Biochem Biophys Res Commun, 2002, 290(3): 998-1009.
doi: 10.1006/bbrc.2001.6299 URL |
| [13] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiol, 2006, 140(2): 411-432.
doi: 10.1104/pp.105.073783 URL |
| [14] |
Dossa K, Wei X, et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress[J]. BMC Plant Biol, 2016, 16(1): 171.
doi: 10.1186/s12870-016-0859-4 URL |
| [15] |
Ito Y, Katsura K, Maruyama K, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice[J]. Plant Cell Physiol, 2006, 47(1): 141-153.
doi: 10.1093/pcp/pci230 URL |
| [16] |
Dubouzet JG, Sakuma Y, Ito Y, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression[J]. Plant J, 2003, 33(4): 751-763.
pmid: 12609047 |
| [17] |
Qin F, Sakuma Y, Li J, et al. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L[J]. Plant Cell Physiol, 2004, 45(8): 1042-1052.
doi: 10.1093/pcp/pch118 URL |
| [18] |
Chen M, Xu Z, et al. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean(Glycine max L.)[J]. J Exp Bot, 2009, 60(1): 121-135.
doi: 10.1093/jxb/ern269 URL |
| [19] |
Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance[J]. J Exp Bot, 2007, 58(2): 221-227.
pmid: 17075077 |
| [20] |
Gilmour SJ, Sebolt AM, Salazar MP, et al. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation[J]. Plant Physiol, 2000, 124(4): 1854-1865.
pmid: 11115899 |
| [21] | Zong JM, Li XW, Zhou YH, et al. The AaDREB1 transcription factor from the cold-tolerant plant Adonis amurensis enhances abiotic stress tolerance in transgenic plant[J]. Int J Mol Sci, 2016, 17(4): E611. |
| [22] | Liu BJ, Zhou Y, Lan W, et al. LlDREB1G, a novel DREB subfamily gene from Lilium longiflorum, can enhance transgenic Arabidopsis tolerance to multiple abiotic stresses[J]. Plant Cell Tissue Organ Cult PCTOC, 2019, 138(3): 489-506. |
| [23] |
Knight H, Zarka DG, Okamoto H, et al. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element[J]. Plant Physiol, 2004, 135(3): 1710-1717.
doi: 10.1104/pp.104.043562 URL |
| [24] |
Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene responsive factor(AP2/ERF)transcription factors:mediators of stress responses and developmental programs[J]. New Phytol, 2013, 199(3): 639-649.
doi: 10.1111/nph.12291 URL |
| [25] |
Liu Y, Zhao TJ, Liu JM, et al. The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box[J]. FEBS Lett, 2006, 580(5): 1303-1308.
doi: 10.1016/j.febslet.2006.01.048 URL |
| [26] | 高文俊, 徐静, 谢开云, 等. Na2CO3和NaHCO3胁迫下冰草的生长及生理响应[J]. 草业学报, 2011, 20(4): 299-304. |
| Gao WJ, Xu J, Xie KY, et al. Physiological responses of Agropyron cristatum under Na2CO3 and NaHCO3 stress[J]. Acta Prataculturae Sin, 2011, 20(4): 299-304. | |
| [27] |
Sakuma Y, Maruyama K, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression[J]. Plant Cell, 2006, 18(5): 1292-1309.
pmid: 16617101 |
| [28] |
Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance[J]. Trends Plant Sci, 2015, 20(4): 219-229.
doi: 10.1016/j.tplants.2015.02.001 URL |
| [29] |
Alam MM, Nahar K, Hasanuzzaman M, et al. Exogenous jasmonic acid modulates the physiology, antioxidant defense and glyoxalase systems in imparting drought stress tolerance in different Brassica species[J]. Plant Biotechnol Rep, 2014, 8(3): 279-293.
doi: 10.1007/s11816-014-0321-8 URL |
| [30] | Yosefi A, Mozafari AA, Javadi T. Jasmonic acid improved in vitro strawberry(Fragaria × ananassa Duch. )resistance to PEG-induced water stress[J]. Plant Cell Tissue Organ Cult PCTOC, 2020, 142(3): 549-558. |
| [31] |
Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J, 2011, 65(6): 907-921.
doi: 10.1111/tpj.2011.65.issue-6 URL |
| [32] |
Cao X, Yan J, Lei J, et al. De novo transcriptome sequencing of MeJA-induced Taraxacum koksaghyz Rodin to identify genes related to rubber formation[J]. Sci Rep, 2017, 7(1): 15697.
doi: 10.1038/s41598-017-14890-z URL |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [3] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [4] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [5] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [6] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [7] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [8] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [9] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [10] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [11] | 刘珍银, 段郅臻, 彭婷, 王童欣, 王健. 基于三角梅的病毒诱导基因沉默体系的建立与优化[J]. 生物技术通报, 2023, 39(7): 123-130. |
| [12] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [13] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [14] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [15] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||