








生物技术通报 ›› 2022, Vol. 38 ›› Issue (11): 238-249.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0167
赵宝顶( ), 吕佳, 申玉玉, 桂玲, 陈钟秀, 陈杰, 路福平, 黎明(
), 吕佳, 申玉玉, 桂玲, 陈钟秀, 陈杰, 路福平, 黎明( )
)
收稿日期:2022-02-15
出版日期:2022-11-26
发布日期:2022-12-01
作者简介:赵宝顶,男,硕士研究生,研究方向:微生物与生化药学;E-mail:基金资助:
ZHAO Bao-ding( ), LV Jia, SHEN Yu-yu, GUI Ling, CHEN Zhong-xiu, CHEN Jie, LU Fu-ping, LI Ming(
), LV Jia, SHEN Yu-yu, GUI Ling, CHEN Zhong-xiu, CHEN Jie, LU Fu-ping, LI Ming( )
)
Received:2022-02-15
Published:2022-11-26
Online:2022-12-01
摘要:
以大肠杆菌作为嘧啶核糖核苷水解酶(pyrimidine-specific ribonucleoside hydrolase RihA,RihA)表达宿主,利用生物转化的方法将尿苷高效转化为尿嘧啶。首先构建pET22b-RihA质粒,在大肠杆菌BL21(DE3)中重组表达,研究尿苷的转化情况。采用优化pET22b-RihA质粒信号肽和与分子伴侣质粒共表达两种策略进一步提高大肠杆菌转化尿苷的效率。来源于碱性磷酸酶的PhoA信号肽和分子伴侣GroES-GroEL共表达的菌株F在投底物发酵时转化效果最好,当底物浓度为65 g/L转化15 h时,菌株F几乎将尿苷完全转化,尿苷转化率达到98.9%,而原始菌株A的尿苷转化率仅为80.2%。进一步对菌株F转化尿苷的浓度进行优化,投入一倍体积的尿苷底物后继续培养约53 h尿苷转化完全,得到尿嘧啶产量为73.45 g/L,尿嘧啶产率为98.16%。发酵液上清中RihA酶活最高的为PelB信号肽和分子伴侣GroES-GroEL共表达的菌株C,其RihA酶活是原始菌株A的10.0倍。其中总可溶性RihA酶活最高的为PhoA信号肽和分子伴侣DnaK-DnaJ-GrpE共表达的菌株G,其RihA酶活是原始菌株A的4.45倍。通过优化信号肽和分子伴侣,提高了尿苷的转化效率,并建立了一种高效的利用大肠杆菌全细胞催化生产药物中间体尿嘧啶的方法。与化学合成方法生产相比,避免了繁琐的生产工艺步骤,对环境友好绿色。
赵宝顶, 吕佳, 申玉玉, 桂玲, 陈钟秀, 陈杰, 路福平, 黎明. 基于信号肽和分子伴侣策略促进大肠杆菌高效转化尿苷[J]. 生物技术通报, 2022, 38(11): 238-249.
ZHAO Bao-ding, LV Jia, SHEN Yu-yu, GUI Ling, CHEN Zhong-xiu, CHEN Jie, LU Fu-ping, LI Ming. Efficient Transformation of Uridine by Escherichia coli Based on Signal Peptide and Molecular Chaperone Strategy[J]. Biotechnology Bulletin, 2022, 38(11): 238-249.
| 菌株和质粒 Strain and plasmid | 相关特性 Related characteristics | 来源 Source | |
|---|---|---|---|
| 菌株 Strain | E. coli BL21 | Wild type E. coli BL21(DE3) | Lab stock |
| E. coli DH5α | Cloning host | Lab stock | |
| E. coli K12. MG1655 | Wild type | Lab stock | |
| SP-1 | BL21 derivate with plasmid pET22b-Fhud-RihA | This work | |
| SP-2 | BL21 derivate with plasmid pET22b-OmpA-RihA | This work | |
| SP-3 | BL21 derivate with plasmid pET22b-OmpC-RihA | This work | |
| SP-4 | BL21 derivate with plasmid pET22b-OmpF-RihA | This work | |
| SP-5 | BL21 derivate with plasmid pET22b-OmpT-RihA | This work | |
| SP-6 | BL21 derivate with plasmid pET22b-PhoE-RihA | This work | |
| BL21-pCDM4-DnaJ | BL21 derivate with plasmid BL21-pCDM4-DnaJ | This work | |
| BL21-pCDM4-DnaK | BL21 derivate with plasmid BL21-pCDM4-DnaK | This work | |
| BL21-pCDM4-GroEL | BL21 derivate with plasmid BL21-pCDM4-GroEL | This work | |
| BL21-pCDM4-GroES | BL21 derivate with plasmid BL21-pCDM4-GroES | This work | |
| BL21-pCDM4-GrpE | BL21 derivate with plasmid BL21-pCDM4-GrpE | This work | |
| Strain A | BL21 derivate with plasmid pET22b-RihA | This work | |
| Strain B | BL21 derivate with plasmid pET22b-PhoA-RihA | This work | |
| Strain C | BL21 derivate with plasmid pET22b-RihA and pCDM4-GroES-GroEL | This work | |
| Strain D | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-DrpE | This work | |
| Strain E | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| Strain F | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-GroES-GroEL | This work | |
| Strain G | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-DnaK-DnaJ-GrpE | This work | |
| Strain H | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| 质粒Plasmid | pET22b | T7 promoters,AmpR | Lab stock |
| pCDM4 | T7 promoters,SmR | [ | |
| pET22b-RihA | pET22b derivate with rihA cloned | This work | |
| pET22b-PhoA-RihA | pET22b derivate with PhoA-rihA cloned | This work | |
| pET22b-Fhud-RihA | pET22b derivate with Fhud-rihA cloned | This work | |
| pET22b-OmpA-RihA | pET22b derivate with OmpA-rihA cloned | This work | |
| pET22b-OmpC-RihA | pET22b derivate with OmpC-rihA cloned | This work | |
| pET22b-OmpF-RihA | pET22b derivate with OmpF-rihA cloned | This work | |
| pET22b-OmpT-RihA | pET22b derivate with OmpT-rihA cloned | This work | |
| pET22bpET22b-PhoE-RihA | pET22b derivate with PhoE-rihA cloned | This work | |
| pCDM4-DnaJ | pCDM4 derivate with dnaJ cloned | This work | |
| pCDM4-DnaK | pCDM4 derivate with dnaK cloned | This work | |
| pCDM4-GroEL | pCDM4 derivate with groEL cloned | This work | |
| pCDM4-GroES | pCDM4 derivate with groES cloned | This work | |
| pCDM4-GrpE | pCDM4 derivate with grpE cloned | This work | |
| pCDM4-GroES-GroEL | pCDM4 derivate with groES-groEL cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE | pCDM4 derivate with dnaK-dnaJ-grpE cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE- GroES-GroEL | pCDM4 derivate with dnaK-dnaJ-grpE-groES-groEL cloned | This work |
表1 本研究所用的菌株及质粒
Table 1 Strains and plasmids in this study
| 菌株和质粒 Strain and plasmid | 相关特性 Related characteristics | 来源 Source | |
|---|---|---|---|
| 菌株 Strain | E. coli BL21 | Wild type E. coli BL21(DE3) | Lab stock |
| E. coli DH5α | Cloning host | Lab stock | |
| E. coli K12. MG1655 | Wild type | Lab stock | |
| SP-1 | BL21 derivate with plasmid pET22b-Fhud-RihA | This work | |
| SP-2 | BL21 derivate with plasmid pET22b-OmpA-RihA | This work | |
| SP-3 | BL21 derivate with plasmid pET22b-OmpC-RihA | This work | |
| SP-4 | BL21 derivate with plasmid pET22b-OmpF-RihA | This work | |
| SP-5 | BL21 derivate with plasmid pET22b-OmpT-RihA | This work | |
| SP-6 | BL21 derivate with plasmid pET22b-PhoE-RihA | This work | |
| BL21-pCDM4-DnaJ | BL21 derivate with plasmid BL21-pCDM4-DnaJ | This work | |
| BL21-pCDM4-DnaK | BL21 derivate with plasmid BL21-pCDM4-DnaK | This work | |
| BL21-pCDM4-GroEL | BL21 derivate with plasmid BL21-pCDM4-GroEL | This work | |
| BL21-pCDM4-GroES | BL21 derivate with plasmid BL21-pCDM4-GroES | This work | |
| BL21-pCDM4-GrpE | BL21 derivate with plasmid BL21-pCDM4-GrpE | This work | |
| Strain A | BL21 derivate with plasmid pET22b-RihA | This work | |
| Strain B | BL21 derivate with plasmid pET22b-PhoA-RihA | This work | |
| Strain C | BL21 derivate with plasmid pET22b-RihA and pCDM4-GroES-GroEL | This work | |
| Strain D | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-DrpE | This work | |
| Strain E | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| Strain F | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-GroES-GroEL | This work | |
| Strain G | BL21 derivate with plasmid pET22b-PhoA-RihA and pCDM4-DnaK-DnaJ-GrpE | This work | |
| Strain H | BL21 derivate with plasmid pET22b-RihA and pCDM4-DnaK-DnaJ-GrpE-GroES-GroEL | This work | |
| 质粒Plasmid | pET22b | T7 promoters,AmpR | Lab stock |
| pCDM4 | T7 promoters,SmR | [ | |
| pET22b-RihA | pET22b derivate with rihA cloned | This work | |
| pET22b-PhoA-RihA | pET22b derivate with PhoA-rihA cloned | This work | |
| pET22b-Fhud-RihA | pET22b derivate with Fhud-rihA cloned | This work | |
| pET22b-OmpA-RihA | pET22b derivate with OmpA-rihA cloned | This work | |
| pET22b-OmpC-RihA | pET22b derivate with OmpC-rihA cloned | This work | |
| pET22b-OmpF-RihA | pET22b derivate with OmpF-rihA cloned | This work | |
| pET22b-OmpT-RihA | pET22b derivate with OmpT-rihA cloned | This work | |
| pET22bpET22b-PhoE-RihA | pET22b derivate with PhoE-rihA cloned | This work | |
| pCDM4-DnaJ | pCDM4 derivate with dnaJ cloned | This work | |
| pCDM4-DnaK | pCDM4 derivate with dnaK cloned | This work | |
| pCDM4-GroEL | pCDM4 derivate with groEL cloned | This work | |
| pCDM4-GroES | pCDM4 derivate with groES cloned | This work | |
| pCDM4-GrpE | pCDM4 derivate with grpE cloned | This work | |
| pCDM4-GroES-GroEL | pCDM4 derivate with groES-groEL cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE | pCDM4 derivate with dnaK-dnaJ-grpE cloned | This work | |
| pCDM4-DnaK-DnaJ-GrpE- GroES-GroEL | pCDM4 derivate with dnaK-dnaJ-grpE-groES-groEL cloned | This work |
| 名称 Name | 引物序列Primer sequence(5'-3') |
|---|---|
| pET22bRihAF | cctcgctgcccagccggcgatggccatgGCACTGCCAATTCTGTTAGATTG |
| pET22bRihAR | tgtcgacggagctcgaattcggatccttaAGCGTAAAATTTCAGACGATCAGCC |
| FhuD-RihA F1 | gatgaataccgcccacgcggcggctattgatcccaatatgGCACTGCCAATTCTGTTAGA |
| FhuD F1 | gactgttaacggcgatggcgctttctccgttgttatggcagatgaataccgcccacgcg |
| FhuD F2 | taagaaggagatatacatatgagcggcttacctcttatttcgcgccgtcgactgttaacggcgatggcg |
| OmpA-RihA F1 | gcactggctggtttcgctaccgtagcgcaggccatgGCACTGCCAATTCTGTTAG |
| OmpA F1 | taagaaggagatatacatatgaaaaagacagctatcgcgattgcagtggcactggctggtttcgc |
| OmpC F1 | taagaaggagatatacatatgaaagttaaagtactgtccctcctggtcccagctctgctggtagcag |
| OmpC-RihA F1 | ccagctctgctggtagcaggcgcagcaaacgctatgGCACTGCCAATTCTGTTAGA |
| OmpF F1 | ttaagaaggagatatacatatgatgaagcgcaatattctggcagtgatcgtccctgctctgttagtagcaggt |
| OmpF-RihA F1 | ccctgctctgttagtagcaggtactgcaaacgctatgGCACTGCCAATTCTGTTAG |
| OmpT F1 | tttaagaaggagatatacatatgcgggcgaaacttctgggaatagtcctgacaacccctattgcga |
| OmpT-RihA F1 | cctgacaacccctattgcgatcagctcttttgctatgGCACTGCCAATTCTGTTAG |
| PhoA F1 | tttaagaaggagatatacatatgaaacaaagcactattgcactggcactcttaccgttactgtttacccctgt |
| PhoA-RihA F1 | cttaccgttactgtttacccctgtgacaaaagccatgGCACTGCCAATTCTGTT |
| PhoE F1 | tttaagaaggagatatacatatgaaaaagagcactctggcattagtggtgatgggcattgtggcatct |
| PhoE-RihA F1 | gatgggcattgtggcatctgcatctgtacaggctatgGCACTGCCAATTCTGTTAG |
| dnaJ F1 | GGAATTCCATatggctaagcaagattattacgaga |
| dnaJ R1 | GGACTAGTttagcgggtcaggtcgtca |
| dnaK F1 | GGAATTCCATatgggtaaaataattggtatcgacctgg |
| dnaK R1 | GGACTAGTttattttttgtctttgacttcttcaaattcagc |
| GroEL F1 | GGAATTCCATatggcagctaaagacgtaaaattcg |
| GroEL R1 | GGACTAGTttacatcatgccgcccatgc |
| GroES F1 | GGAATTCCATatgaatattcgtccattgcatgatcg |
| GroES R1 | GGACTAGTttacgcttcaacaattgccagaatgt |
| grpE F1 | GGAATTCCATatgagtagtaaagaacagaaaacgcctg |
| grpE R1 | GGACTAGTttaagcttttgctttcgctacagtaacc |
| Cht F1 | cttgaggggttttttTctagAatcgagatcgatctcgatcccg |
| Cht R1 | actaccggaagcagtgtcgactc |
| To F1 | gatgcgtccggcgtagc |
表2 引物列表
Table 2 List of primers
| 名称 Name | 引物序列Primer sequence(5'-3') |
|---|---|
| pET22bRihAF | cctcgctgcccagccggcgatggccatgGCACTGCCAATTCTGTTAGATTG |
| pET22bRihAR | tgtcgacggagctcgaattcggatccttaAGCGTAAAATTTCAGACGATCAGCC |
| FhuD-RihA F1 | gatgaataccgcccacgcggcggctattgatcccaatatgGCACTGCCAATTCTGTTAGA |
| FhuD F1 | gactgttaacggcgatggcgctttctccgttgttatggcagatgaataccgcccacgcg |
| FhuD F2 | taagaaggagatatacatatgagcggcttacctcttatttcgcgccgtcgactgttaacggcgatggcg |
| OmpA-RihA F1 | gcactggctggtttcgctaccgtagcgcaggccatgGCACTGCCAATTCTGTTAG |
| OmpA F1 | taagaaggagatatacatatgaaaaagacagctatcgcgattgcagtggcactggctggtttcgc |
| OmpC F1 | taagaaggagatatacatatgaaagttaaagtactgtccctcctggtcccagctctgctggtagcag |
| OmpC-RihA F1 | ccagctctgctggtagcaggcgcagcaaacgctatgGCACTGCCAATTCTGTTAGA |
| OmpF F1 | ttaagaaggagatatacatatgatgaagcgcaatattctggcagtgatcgtccctgctctgttagtagcaggt |
| OmpF-RihA F1 | ccctgctctgttagtagcaggtactgcaaacgctatgGCACTGCCAATTCTGTTAG |
| OmpT F1 | tttaagaaggagatatacatatgcgggcgaaacttctgggaatagtcctgacaacccctattgcga |
| OmpT-RihA F1 | cctgacaacccctattgcgatcagctcttttgctatgGCACTGCCAATTCTGTTAG |
| PhoA F1 | tttaagaaggagatatacatatgaaacaaagcactattgcactggcactcttaccgttactgtttacccctgt |
| PhoA-RihA F1 | cttaccgttactgtttacccctgtgacaaaagccatgGCACTGCCAATTCTGTT |
| PhoE F1 | tttaagaaggagatatacatatgaaaaagagcactctggcattagtggtgatgggcattgtggcatct |
| PhoE-RihA F1 | gatgggcattgtggcatctgcatctgtacaggctatgGCACTGCCAATTCTGTTAG |
| dnaJ F1 | GGAATTCCATatggctaagcaagattattacgaga |
| dnaJ R1 | GGACTAGTttagcgggtcaggtcgtca |
| dnaK F1 | GGAATTCCATatgggtaaaataattggtatcgacctgg |
| dnaK R1 | GGACTAGTttattttttgtctttgacttcttcaaattcagc |
| GroEL F1 | GGAATTCCATatggcagctaaagacgtaaaattcg |
| GroEL R1 | GGACTAGTttacatcatgccgcccatgc |
| GroES F1 | GGAATTCCATatgaatattcgtccattgcatgatcg |
| GroES R1 | GGACTAGTttacgcttcaacaattgccagaatgt |
| grpE F1 | GGAATTCCATatgagtagtaaagaacagaaaacgcctg |
| grpE R1 | GGACTAGTttaagcttttgctttcgctacagtaacc |
| Cht F1 | cttgaggggttttttTctagAatcgagatcgatctcgatcccg |
| Cht R1 | actaccggaagcagtgtcgactc |
| To F1 | gatgcgtccggcgtagc |
| 信号肽 Signal peptide | 来源 Source | 氨基酸序列 Amino acid sequence |
|---|---|---|
| Fhud | Iron(III)hydroxamate ABC transporter periplasmic binding protein | MSGLPLISRRRLLTAMALSPLLWQMNTAHAAAIDPN |
| OmpA | Outer-membrane protein A | MKKTAIAIAVALAGFATVAQA |
| OmpC | Outer-membrane protein C | MKVKVLSLLVPALLVAGAANA |
| OmpF | Outer-membrane protein F | MMKRNILAVIVPALLVAGTANA |
| OmpT | Protease 7 | MRAKLLGIVLTTPIAISSFA |
| PhoA | Alkaline phosphatase | VKQSTIALALLPLLFTPVTKA |
| PhoE | Outer-membrane pore protein F | MKKSTLALVVMGIVASASVQA |
| PelB | Pectin lyase | MKYLLPTAAAGLLLLAAQPAMA |
表3 信号肽的序列及来源
Table 3 Sequences and sources of signal peptides
| 信号肽 Signal peptide | 来源 Source | 氨基酸序列 Amino acid sequence |
|---|---|---|
| Fhud | Iron(III)hydroxamate ABC transporter periplasmic binding protein | MSGLPLISRRRLLTAMALSPLLWQMNTAHAAAIDPN |
| OmpA | Outer-membrane protein A | MKKTAIAIAVALAGFATVAQA |
| OmpC | Outer-membrane protein C | MKVKVLSLLVPALLVAGAANA |
| OmpF | Outer-membrane protein F | MMKRNILAVIVPALLVAGTANA |
| OmpT | Protease 7 | MRAKLLGIVLTTPIAISSFA |
| PhoA | Alkaline phosphatase | VKQSTIALALLPLLFTPVTKA |
| PhoE | Outer-membrane pore protein F | MKKSTLALVVMGIVASASVQA |
| PelB | Pectin lyase | MKYLLPTAAAGLLLLAAQPAMA |
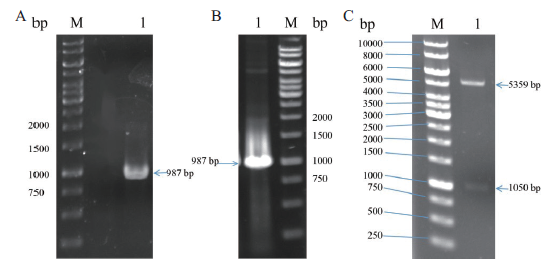
图3 pET22b-RihA质粒的构建及验证 A:rihA基因PCR扩增(M:DNA marke,1:rihA);B:重组质粒pET22b-RihA的菌落PCR验证(M:DNA marker,1:rihA);C:重组质粒pET22b-RihA双酶切验证(M:DNA marker,1:pET22b-RihA)
Fig. 3 Construction and validation of pET22b-RihA plasmid A:PCR amplification product of rihA(M:DNA marker,1:rihA). B:Colony PCR validation of recombinant plasmid pET22b-RihA(M:DNA marker,1:rih A). C:Double digestion verification of recombinant plasmid pET22b-RihA(M:DNA marker,1:pET22b-RihA)
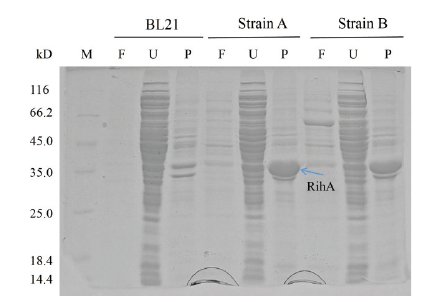
图4 菌株A、菌株B和BL21菌株表达产物的SDS-PAGE分析 M:蛋白标准品;F:发酵液上清;U:超声破碎上清;P:超声破碎沉淀
Fig.4 SDS-PAGE analysis of expression products of strain A,B and BL21 M:Molecular weight marker;F:fermentation supernatant;U:ultrasonication supernatant;P:precipitation
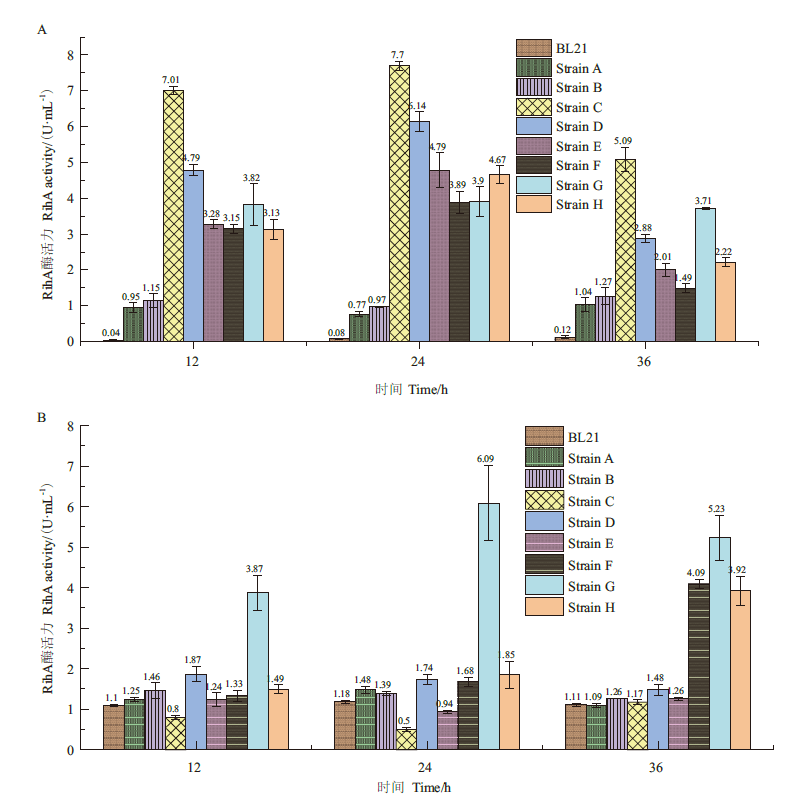
图5 9株菌在不同时间胞外和胞内RihA酶活力 A:发酵液上清中RihA酶活力;B:菌体超声破碎上清RihA酶活力
Fig.8 Extracellular and intracellular RihA activities of 9 strains at different time A:Extracellular RihA enzyme activity. B:Intracellular RihA enzyme activity
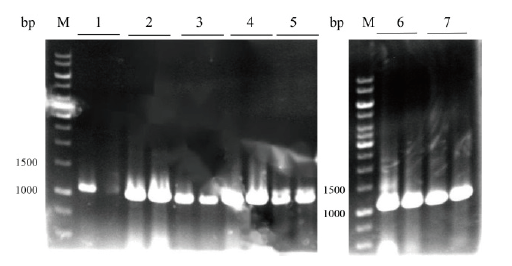
图6 7种信号肽质粒pET22b-Fhud/OmpA/OmpC/OmpF/OmpT/PhoA/PhoE-RihA 菌落PCR验证
Fig.6 Colony PCR verification of seven kinds of signal pep-tide plasmid pET22b-Fhud/OmpA/OmpC/OmpF/OmpT/PhoA/PhoE-RihA M:DNA marker;1:Fhud-rihA(1 088 bp);2:OmpA-rihA(1 043 bp);3:OmpC-rihA(1 043 bp);4:OmpF-rihA(1 047 bp);5:OmpT-rihA(1 042 bp);6:PhoA-rihA(1 045 bp);7:PhoE-rihA(1 045 bp)
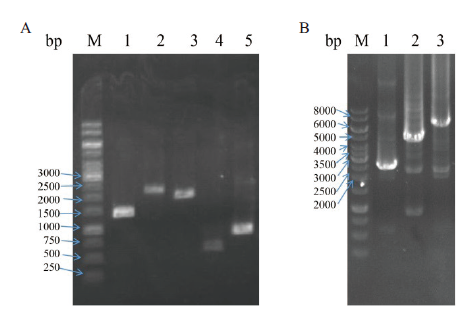
图8 串联分子伴侣质粒的构建及验证 A:不同分子伴侣表达盒的PCR扩增,M:DNA marker,1:T7P-dnaJ-T7T(1 401 bp),2:T7P-dnaK-T7T(2 187 bp),3:T7P-groEL-T7T(1 917 bp),4:T7P-groES-T7T(564 bp),5:T7P-grpE-T7T(864 bp);B:串联分子伴侣质粒的PCR验证,M:DNA marker,1:groES-groEL(2 429 bp),2:dnaK-dnaJ-grpE(4 348 bp),3:dnaK-dnaJ-grpE-groES-groEL(6 725 bp)
Fig.8 Construction and validation of tandem molecular chaperone plasmids A:PCR amplification of different chaperone expression boxes,M:DNA marker,1:T7P-dnaJ-T7T(1 401 bp),2:T7P-dnaK-T7T(2 187 bp),3:T7P-groEL-T7T(1 917 bp),4:T7P-groES-T7T(564 bp),5:T7P-grpE-T7T(864 bp). B:PCR validation of tandem molecular chaperone plasmids,M:DNA marker,1:groES-groEL(2 429 bp),2:dnaK-dnaJ-grpE(4 348 bp),3:dnaK-dnaJ-grpE-groES-groEL(6 725 bp)

图9 不同菌株表达产物的SDS-PAGE分析 M:蛋白标准品;F:发酵液上清;U:超声破碎上清;P:超声破碎沉淀
Fig.9 SDS-PAGE analysis of expression products of differ-ent strains M:Molecular weight marker;F:fermentation supernatant;U:ultrasonication supernatant;P:precipitation
| 投底物量 Amount of the substrate | 发酵液中初始尿苷浓度 Initial uridine concentration of fermentation broth/(g·L-1) | 尿嘧啶产量 Uracil yield/(g·L-1) | 尿嘧啶产率 Uracil productive rate /% |
|---|---|---|---|
| 1倍体积/20 mL One time the volume/20 mL | 163.02±4.50 | 73.45±1.03 | 98.16±1.38 |
| 1.5倍体积/30 mL 1.5 times the volume/30 mL | 195.60±6.01 | 88.09±1.33 | 98.12±1.48 |
| 2倍体积/40 mL Two times the volume/40 mL | 218.78±3.95 | 97.90±2.17 | 97.49±2.16 |
表4 不同底物浓度转化时尿嘧啶产量及产率
Table 4 Uracil yield and uracil productive rate transformed with different substrate concentration
| 投底物量 Amount of the substrate | 发酵液中初始尿苷浓度 Initial uridine concentration of fermentation broth/(g·L-1) | 尿嘧啶产量 Uracil yield/(g·L-1) | 尿嘧啶产率 Uracil productive rate /% |
|---|---|---|---|
| 1倍体积/20 mL One time the volume/20 mL | 163.02±4.50 | 73.45±1.03 | 98.16±1.38 |
| 1.5倍体积/30 mL 1.5 times the volume/30 mL | 195.60±6.01 | 88.09±1.33 | 98.12±1.48 |
| 2倍体积/40 mL Two times the volume/40 mL | 218.78±3.95 | 97.90±2.17 | 97.49±2.16 |
| [1] |
Ganyecz Á, Kállay M, Csontos J. Thermochemistry of uracil, thymine, cytosine, and adenine[J]. J Phys Chem A, 2019, 123(18):4057-4067.
doi: 10.1021/acs.jpca.9b02061 pmid: 30977653 |
| [2] | Ramesh D, Vijayakumar BG, Kannan T. Therapeutic potential of uracil and its derivatives in countering pathogenic and physiological disorders[J]. Eur J Med Chem, 2020, 207:112801. |
| [3] |
Pałasz A, Cież D. In search of uracil derivatives as bioactive agents. Uracils and fused uracils:Synthesis, biological activity and applications[J]. Eur J Med Chem, 2015, 97:582-611.
doi: 10.1016/j.ejmech.2014.10.008 URL |
| [4] | 王利敏, 王思瑶, 张诗缇, 等. 医药中间体尿嘧啶的合成研究[J]. 辽宁医学院学报, 2014, 35(6):7-9. |
| Wang LM, Wang SY, Zhang ST, et al. Study on the synthesis of uracil as a pharmaceutical intermediate[J]. J Liaoning Med Univ, 2014, 35(6):7-9. | |
| [5] |
Kim S, Lee WJ, Song I, et al. Production of uracil from methane by a newly isolated Methylomonas sp. SW1[J]. J Biotechnol, 2016, 240:43-47.
doi: 10.1016/j.jbiotec.2016.10.019 URL |
| [6] |
Lee WJ, Kim S, Song I, et al. Microbial production of uracil by an isolated Methylobacterium sp. WJ4 using methanol[J]. Enzyme Microb Technol, 2018, 111:63-66.
doi: 10.1016/j.enzmictec.2017.10.003 URL |
| [7] |
Petersen C, Møller LB. The RihA RihB, and RihC ribonucleoside hydrolases of Escherichia coli. substrate specificity, gene expression, and regulation[J]. J Biol Chem, 2001, 276(2):884-894.
doi: 10.1074/jbc.M008300200 pmid: 11027694 |
| [8] |
Versées W, Steyaert J. Catalysis by nucleoside hydrolases[J]. Curr Opin Struct Biol, 2003, 13(6):731-738.
doi: 10.1016/j.sbi.2003.10.002 URL |
| [9] |
Yoon SH, Kim SK, Kim JF. Secretory production of recombinant proteins in Escherichia coli[J]. Recent Pat Biotechnol, 2010, 4(1):23-29.
doi: 10.2174/187220810790069550 URL |
| [10] | Mergulhão FJ, Monteiro GA. Periplasmic targeting of recombinant proteins in Escherichia coli[M]// van der Giezen M. Protein Targeting Protocols. Totowa, NJ: Humana Press, 2007:47-61. |
| [11] | Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli:advances and challenges[J]. Front Microbiol, 2014, 5:172. |
| [12] |
Singhvi P, Saneja A, Srichandan S, et al. Bacterial inclusion bodies:a treasure trove of bioactive proteins[J]. Trends Biotechnol, 2020, 38(5):474-486.
doi: 10.1016/j.tibtech.2019.12.011 URL |
| [13] |
Kleiner-Grote GRM, Risse JM, Friehs K. Secretion of recombinant proteins from E. coli[J]. Eng Life Sci, 2018, 18(8):532-550.
doi: 10.1002/elsc.201700200 pmid: 32624934 |
| [14] |
Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli[J]. Nat Biotechnol, 2004, 22(11):1399-1408.
doi: 10.1038/nbt1029 URL |
| [15] |
Oganesyan N, Ankoudinova I, Kim SH, et al. Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization[J]. Protein Expr Purif, 2007, 52(2):280-285.
doi: 10.1016/j.pep.2006.09.015 URL |
| [16] |
Fatima K, Naqvi F, Younas H. A review:molecular chaperone-mediated folding, unfolding and disaggregation of expressed recombinant proteins[J]. Cell Biochem Biophys, 2021, 79(2):153-174.
doi: 10.1007/s12013-021-00970-5 pmid: 33634426 |
| [17] |
Tian YX, Chen J, Yu HM, et al. Overproduction of the Escherichia coli chaperones GroEL-GroES in Rhodococcus ruber improves the activity and stability of cell catalysts harboring a nitrile hydratase[J]. J Microbiol Biotechnol, 2016, 26(2):337-346.
doi: 10.4014/jmb.1509.09084 URL |
| [18] | Yao D, Fan J, Han RZ, et al. Enhancing soluble expression of sucrose phosphorylase in Escherichia coli by molecular chaperones[J]. Protein Expr Purif, 2020, 169:105571. |
| [19] |
Pan D, Zha X, Yu XH, et al. Enhanced expression of soluble human papillomavirus L1 through coexpression of molecular chaperonin in Escherichia coli[J]. Protein Expr Purif, 2016, 120:92-98.
doi: 10.1016/j.pep.2015.12.016 URL |
| [20] | 邓通, 周海胜, 吴坚平, 等. 基于分子伴侣策略提高NADPH依赖型醇脱氢酶的异源可溶性表达[J]. 中国生物工程杂志, 2020, 40(8):24-32. |
| Deng T, Zhou HS, Wu JP, et al. Enhance soluble heteroexpression of a NADPH-dependent alcohol dehydrogenase based on the chaperone strategy[J]. China Biotechnol, 2020, 40(8):24-32. | |
| [21] |
Low KO, Muhammad Mahadi N, Md Illias R. Optimisation of signal peptide for recombinant protein secretion in bacterial hosts[J]. Appl Microbiol Biotechnol, 2013, 97(9):3811-3826.
doi: 10.1007/s00253-013-4831-z pmid: 23529680 |
| [22] |
Mirzadeh K, Shilling PJ, Elfageih R, et al. Increased production of periplasmic proteins in Escherichia coli by directed evolution of the translation initiation region[J]. Microb Cell Fact, 2020, 19(1):85.
doi: 10.1186/s12934-020-01339-8 pmid: 32264894 |
| [23] |
Brockmeier U, Caspers M, Freudl R, et al. Systematic screening of all signal peptides from Bacillus subtilis:a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria[J]. J Mol Biol, 2006, 362(3):393-402.
pmid: 16930615 |
| [24] |
Mathiesen G, Sveen A, Brurberg MB, et al. Genome-wide analysis of signal peptide functionality in Lactobacillus plantarum WCFS1[J]. BMC Genomics, 2009, 10:425.
doi: 10.1186/1471-2164-10-425 pmid: 19744343 |
| [25] |
Freudl R. Signal peptides for recombinant protein secretion in bacterial expression systems[J]. Microb Cell Fact, 2018, 17(1):52.
doi: 10.1186/s12934-018-0901-3 pmid: 29598818 |
| [26] |
Xu P, Vansiri A, Bhan N, et al. ePathBrick:a synthetic biology platform for engineering metabolic pathways in E. coli[J]. ACS Synth Biol, 2012, 1(7):256-266.
doi: 10.1021/sb300016b URL |
| [27] |
Freilich R, Arhar T, Abrams JL, et al. Protein-protein interactions in the molecular chaperone network[J]. Acc Chem Res, 2018, 51(4):940-949.
doi: 10.1021/acs.accounts.8b00036 URL |
| [28] |
Tang YC, Chang HC, Roeben A, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein[J]. Cell, 2006, 125(5):903-914.
doi: 10.1016/j.cell.2006.04.027 URL |
| [29] |
Clare DK, Bakkes PJ, van Heerikhuizen H, et al. Chaperonin complex with a newly folded protein encapsulated in the folding chamber[J]. Nature, 2009, 457(7225):107-110.
doi: 10.1038/nature07479 URL |
| [30] |
Mayer MP, Bukau B. Hsp70 chaperones:cellular functions and molecular mechanism[J]. Cell Mol Life Sci, 2005, 62(6):670-684.
pmid: 15770419 |
| [31] |
Deuerling E, Schulze-Specking A, Tomoyasu T, et al. Trigger factor and DnaK cooperate in folding of newly synthesized proteins[J]. Nature, 1999, 400(6745):693-696.
doi: 10.1038/23301 URL |
| [32] |
Genevaux P, Keppel F, Schwager F, et al. In vivo analysis of the overlapping functions of DnaK and trigger factor[J]. EMBO Rep, 2004, 5(2):195-200.
pmid: 14726952 |
| [33] |
Garavito MF, Narváez-Ortiz HY, Zimmermann BH. Pyrimidine metabolism:dynamic and versatile pathways in pathogens and cellular development[J]. Journal of genetics and genomics, 2015, 42(5):195-205.
doi: 10.1016/j.jgg.2015.04.004 pmid: 26059768 |
|
Garavito MF, Narváez-Ortiz HY, Zimmermann BH. Pyrimidine metabolism:dynamic and versatile pathways in pathogens and cellular development[J]. J Genet Genomics, 2015, 42(5):195-205.
doi: 10.1016/j.jgg.2015.04.004 pmid: 26059768 |
|
| [34] |
West TP. Isolation and characterization of an Escherichia coli B mutant strain defective in uracil catabolism[J]. Can J Microbiol, 1998, 44(11):1106-1109.
pmid: 10030006 |
| [1] | 梅欢, 李玥, 刘可蒙, 刘吉华. 小檗碱桥酶高效原核表达及生物合成l-SLR的研究[J]. 生物技术通报, 2023, 39(7): 277-287. |
| [2] | 董聪, 高庆华, 王玥, 罗同阳, 王庆庆. 基于联合策略提高FAD依赖的葡萄糖脱氢酶的酵母表达[J]. 生物技术通报, 2023, 39(6): 316-324. |
| [3] | 陈广霞, 李秀杰, 蒋锡龙, 单雷, 张志昌, 李勃. 植物小分子信号肽参与非生物逆境胁迫应答的研究进展[J]. 生物技术通报, 2023, 39(11): 61-73. |
| [4] | 段绪果, 张玉华, 黄婷婷, 丁乾, 栾舒越, 朱秋雨. 化学分子伴侣及诱导条件协同强化Thermotoga maritima α-葡聚糖磷酸化酶可溶性表达[J]. 生物技术通报, 2021, 37(8): 233-242. |
| [5] | 贺小丽, 郭磊周, 韩佳慧, 唐殷, 袁媛, 代其林, 平淑珍, 江世杰. 细菌周质分子伴侣LolA研究进展[J]. 生物技术通报, 2021, 37(8): 275-283. |
| [6] | 苗华彪, 曹艳, 杨梦瀚, 黄遵锡. 基于信号肽策略提高外源蛋白在枯草芽孢杆菌中的表达[J]. 生物技术通报, 2021, 37(6): 259-271. |
| [7] | 闵琪, 高子涵, 姚银, 张华山, 熊海容, 张莉. 共表达HAC1和分子伴侣基因对甘露聚糖酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2020, 36(5): 159-168. |
| [8] | 孙腾, 徐刘佳, 郑明明. 硫辛酸衍生物的制备与活性研究进展[J]. 生物技术通报, 2020, 36(4): 41-46. |
| [9] | 梁昕鑫, 唐丹, 霍毅欣. 蛋白源生物质的绿色生物转化[J]. 生物技术通报, 2020, 36(12): 216-228. |
| [10] | 张钰文, 袁航, 于江悦, 马晓晓, 史超硕, 李玉. 一株高效降解羽毛废弃物菌株的筛选及表达条件优化[J]. 生物技术通报, 2019, 35(9): 93-98. |
| [11] | 郭磊周, 韩佳慧, 唐殷, 李江, 黄程, 代其林, 王劲, 平淑珍, 江世杰. DrwH类信号肽序列对其抗氧化功能的影响[J]. 生物技术通报, 2019, 35(5): 125-132. |
| [12] | 赵祥杰, 杨文君, 杨荣玲, 吴婷婷, 王朝宇, 许宁宁, 何佳美. 花色苷生物转化修饰的研究进展[J]. 生物技术通报, 2019, 35(10): 205-211. |
| [13] | 高庆华, 董聪, 王玥, 胡美荣, 王庆庆, 王云鹏, 罗同阳, 刘蕾. 共表达分子伴侣PDI和Ero1对葡萄糖氧化酶在毕赤酵母中表达的影响[J]. 生物技术通报, 2018, 34(7): 174-179. |
| [14] | 杨何宝 ,胡美荣 ,郑翔 ,牟庆璇 ,高沛汝. 不同信号肽及分子伴侣对中性蛋白酶在枯草芽孢杆菌中分泌表达的影响[J]. 生物技术通报, 2018, 34(6): 134-140. |
| [15] | 张亨, 刘盈盈, 陈云, 平淑珍, 王劲. 戈壁异常球菌Dgl5蛋白生物学功能研究[J]. 生物技术通报, 2018, 34(3): 177-184. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||