








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 219-237.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1195
胡永波1,2( ), 雷雨田1,2, 杨永森1,2, 陈馨1,2, 林黄昉1,2, 林碧英1,2, 刘爽1,2, 毕格1,2, 申宝营1,2(
), 雷雨田1,2, 杨永森1,2, 陈馨1,2, 林黄昉1,2, 林碧英1,2, 刘爽1,2, 毕格1,2, 申宝营1,2( )
)
收稿日期:2023-12-15
出版日期:2024-06-26
发布日期:2024-05-14
通讯作者:
申宝营,男,博士,讲师,研究方向:设施园艺;E-mail: shenby889@foxmail.com作者简介:胡永波,男,硕士研究生,研究方向:蔬菜生理生化与生态;E-mail: 194997643@qq.com
基金资助:
HU Yong-bo1,2( ), LEI Yu-tian1,2, YANG Yong-sen1,2, CHEN Xin1,2, LIN Huang-fang1,2, LIN Bi-ying1,2, LIU Shuang1,2, BI Ge1,2, SHEN Bao-ying1,2(
), LEI Yu-tian1,2, YANG Yong-sen1,2, CHEN Xin1,2, LIN Huang-fang1,2, LIN Bi-ying1,2, LIU Shuang1,2, BI Ge1,2, SHEN Bao-ying1,2( )
)
Received:2023-12-15
Published:2024-06-26
Online:2024-05-14
摘要:
【目的】 分析Bcl-2相关抗凋亡(Bcl-2 associated athanogene, BAG)家族蛋白成员在黄瓜和南瓜中响应非生物胁迫以及在黄瓜/南瓜嫁接愈合过程中响应光照的表达模式,为解析黄瓜/南瓜嫁接苗嫁接愈合机理及黄瓜和南瓜等蔬菜的抗性分子育种提供有利基因。【方法】 基于黄瓜和南瓜基因组信息,利用生物信息学手段,对黄瓜和南瓜中BAG基因家族进行鉴定,并对其理化特性、染色体定位、基因结构、系统发育和共线性进行了分析。基于公共数据库及黄瓜/南瓜嫁接苗在嫁接愈合过程中转录组测序数据,分析BAG基因在黄瓜和南瓜中响应非生物胁迫以及在嫁接愈合过程中响应光照的表达模式。【结果】 在黄瓜和南瓜中分别鉴定到12和18个BAG家族基因,均分为2个亚族,基因成员保守性高,I亚家族的BAG主要参与基因调控和逆境响应,而II亚家族的BAG主要参与植物的发育过程。黄瓜和南瓜BAG基因分别与拟南芥、水稻、番茄存在多种线性关系,但CsaV3_1G017210与拟南芥、水稻、番茄和南瓜中的BAG基因均不存在线性关系。不同BAG基因具有组织特异性表达模式。CsaV3_6G000970和CmoCh08G008520(BAG family molecular chaperone regulator 6)在黄瓜和南瓜响应非生物胁迫以及黄瓜/南瓜嫁接苗嫁接愈合过程中均发生上调表达。【结论】 BAG家族基因在黄瓜和南瓜对非生物胁迫的响应以及黄瓜/南瓜嫁接苗嫁接愈合过程中对光的响应中具有差异性,协同调控了黄瓜和南瓜的生长发育及嫁接愈合,在黄瓜和南瓜非生物胁迫以及黄瓜/南瓜嫁接苗嫁接愈合过程中发挥着重要作用。
胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237.
HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch.[J]. Biotechnology Bulletin, 2024, 40(6): 219-237.
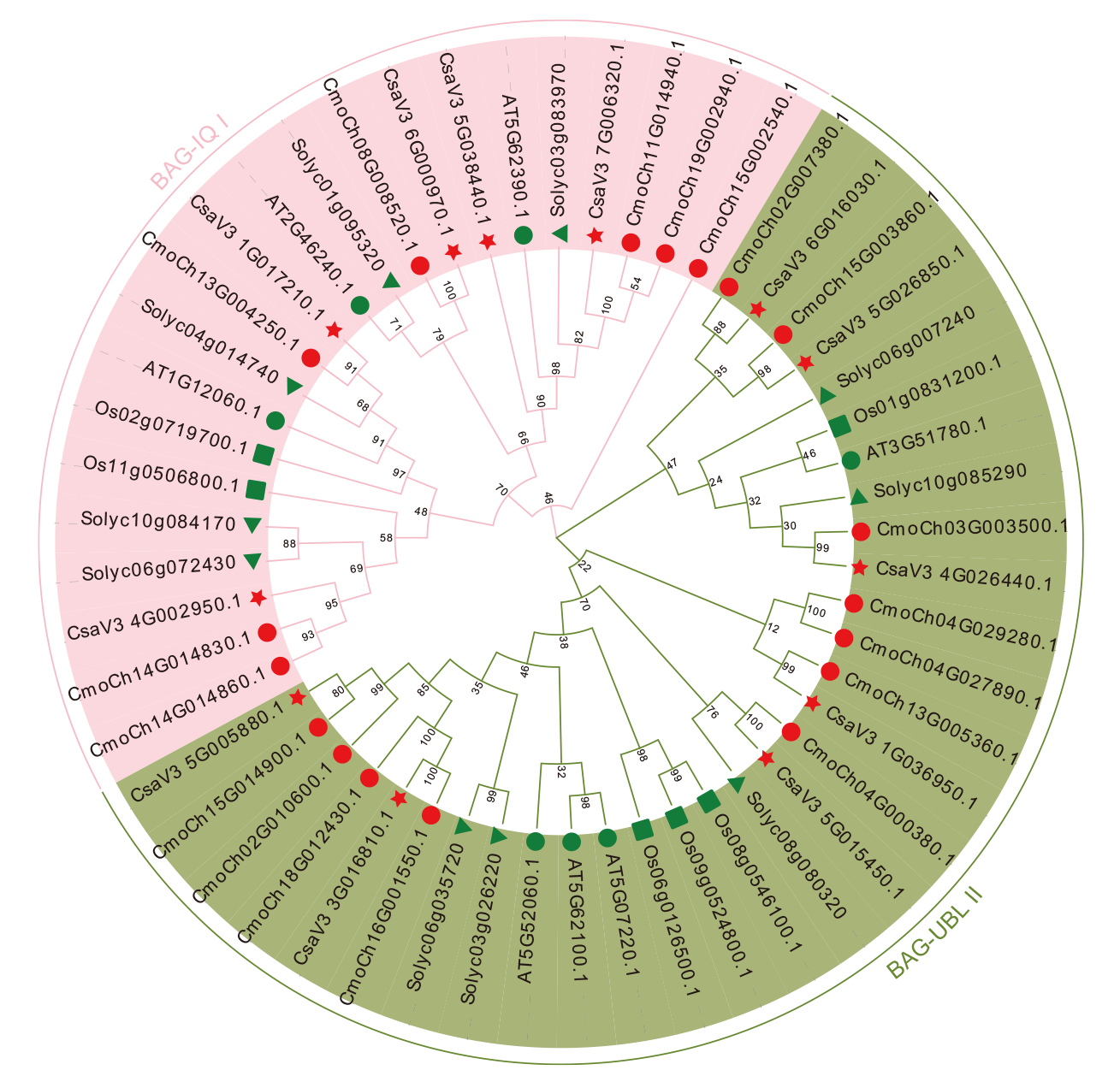
图2 黄瓜、南瓜、拟南芥、水稻和番茄BAG家族基因系统发育分析 :黄瓜; :南瓜; :拟南芥; :水稻; :番茄
Fig. 2 Phylogenetic analysis of the BAG gene family in C. sativus L., C. moschata Duch., Arabidopsis, O. sativa L. and S. lyco-persicum L. : C. sativus L.. : C. moschata Duch.. : Arabidopsis. : O. sativa L.. : S. lycopersicum L

图4 BAG家族基因在黄瓜、南瓜、拟南芥、水稻和番茄中的共线性
Fig. 4 Collinear relationship of BAG gene family in C. sativus L., C. moschata Duch., Arabidopsis, O. sativa L. and S. lycop-ersicum L.

图5 黄瓜(A)和南瓜(B)BAG家族基因启动子序列顺式作用元件分析
Fig. 5 Cis-acting elements in the promoter sequences of BAG gene family in C. sativus L. (A) and C. moschata Duch. (B)
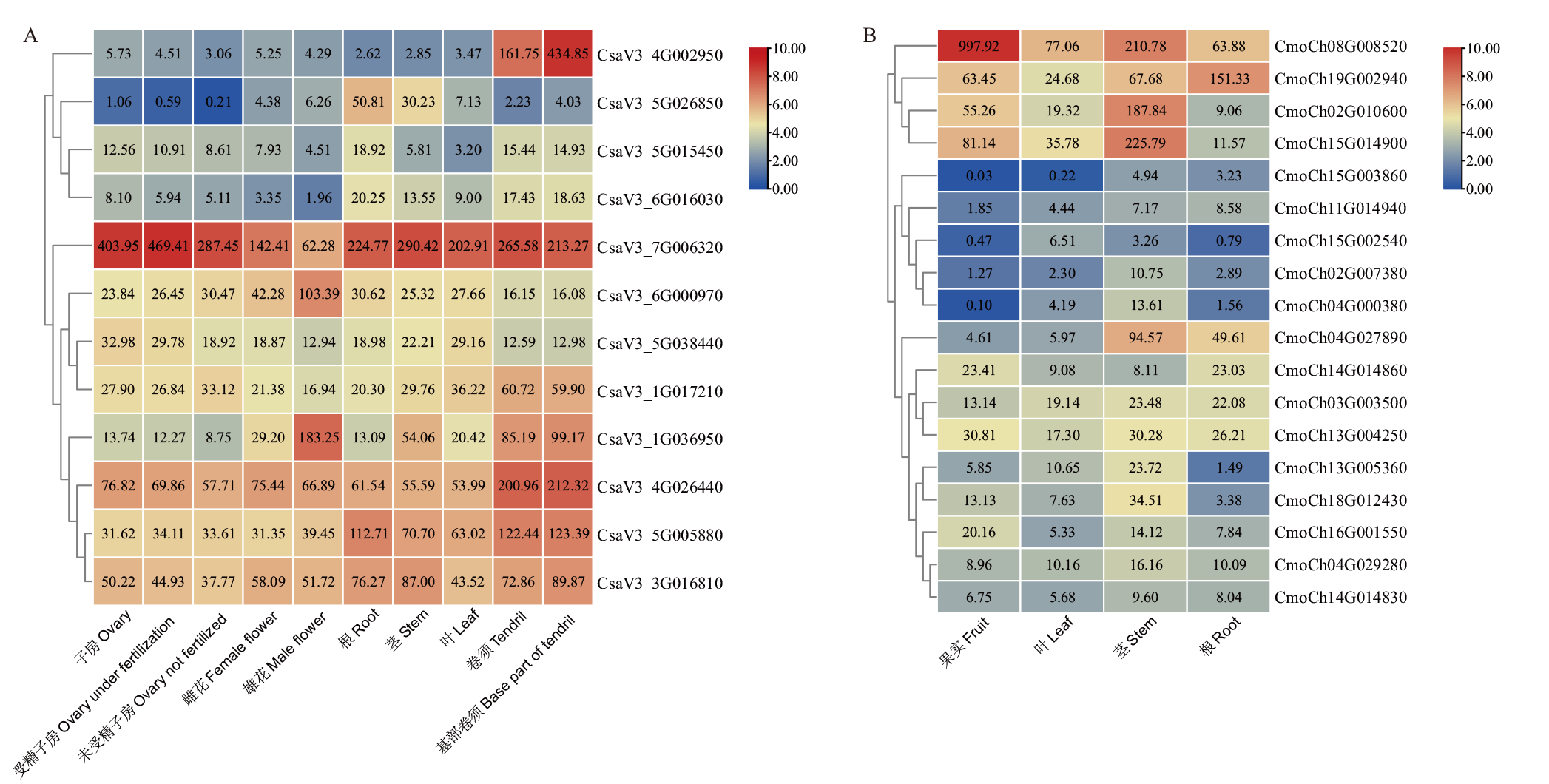
图6 BAG家族基因在黄瓜(A)和南瓜(B)不同组织器官中的表达模式
Fig. 6 Expressions of BAG gene family in different tissues and organs of in C. sativus L. (A) and C. moschata Duch. (B)
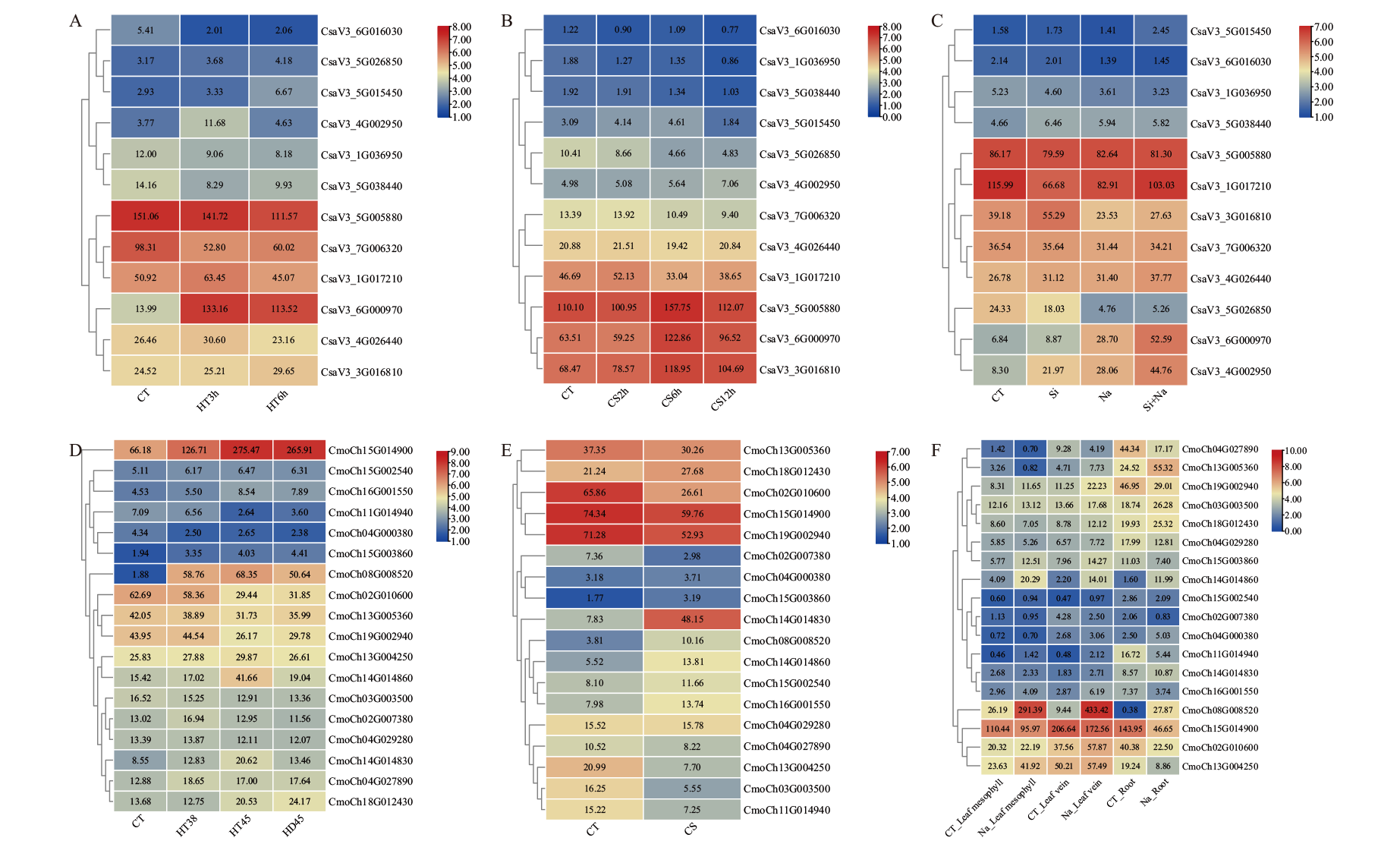
图7 黄瓜和南瓜BAG家族基因在非生物胁迫处理下的表达模式 A:黄瓜高温胁迫处理(HT3h、HT6h:高温处理3、6 h);B:黄瓜低温胁迫处理(CS2h、CS6h、CS12h:低温处理2、6、12 h);C:黄瓜盐和硅胁迫处理(Si:硅胁迫处理;Na:盐胁迫处理;Si+Na:盐和硅胁迫处理);D:南瓜高温胁迫处理(HT38:38℃高温处理3 h;HD45:38℃高温适应3 h,25℃恢复5 h,然后45℃激发3 h;HT45:45℃高温处理3 h);E:南瓜低温胁迫处理(CS:低温处理);F:南瓜盐胁迫处理(CT_Leaf mesophyll、CT_Leaf vein、CT_Root:叶肉、叶脉、根的对照处理;Na_Leaf mesophyll、Na_Leaf vein、Na_Root:叶肉、叶脉、根的盐胁迫处理);CT:对照;表格中的数据是原始的FPKM值
Fig. 7 Expressions of BAG gene family in C. sativus L. and C. moschata Duch. under abiotic stress A: High temperature stress in C. sativus L. (HT3h, HT6h: High temperature treatment for 3 and 6 h). B: Low temperature stress in C. sativus L. (CS2h, CS6h and CS12h: Low temperature treatment for 2, 6 and 12 h). C: Salt and silicon stress in C. sativus L. (Na: Salt stress treatment; Si: silicon stress treatment; Si+Na: silicon and salt stress treatment). D: High temperature stress in C. moschata Duch. (HT38: 38°C high temperature treatment for 3 h; HD45: 38°C high temperature adaptation for 3 h, recovered at 25°C for 5 h, and then excited at 45°C for 3 h; HT45: high temperature treatment at 45°C for 3 h). E: Low temperature stress in C. moschata Duch. (CS: Low temperature treatment). F: Salt stress in C. moschata Duch. (CT_Leaf mesophyll, CT_Leaf vein, and CT_Root: Control treatment of mesophyll, leaf veins, and roots; Na_Leaf mesophyll, Na_Leaf vein and, Na_Root: salt stress treatment of mesophyll, leaf veins, and roots. CT: Control. The data in the boxes indicate original FPKM value
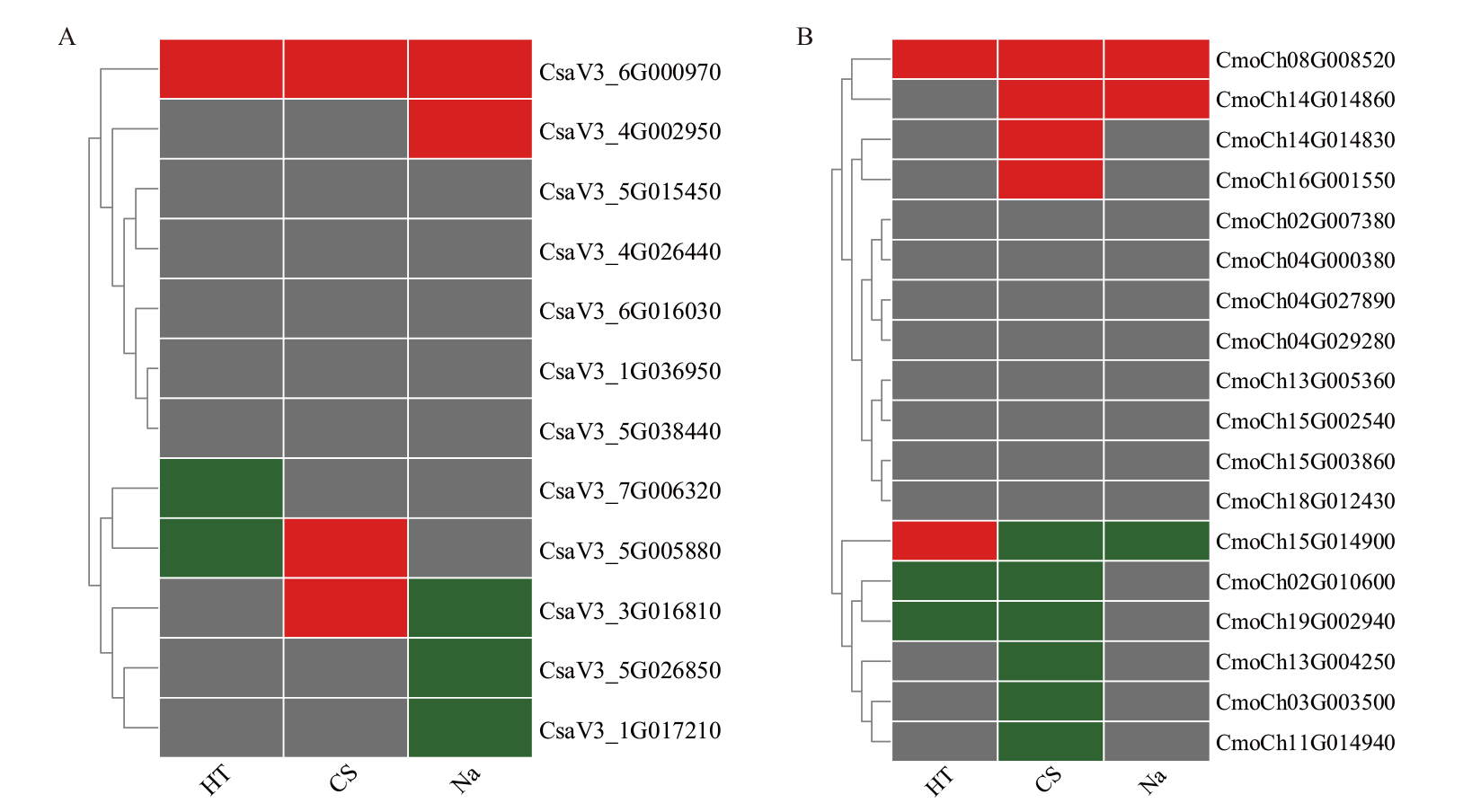
图8 黄瓜(A)和南瓜(B)BAG家族基因在胁迫下的表达模式热图 HT:高温胁迫;CS:低温胁迫;Na:盐胁迫。其中,红色代表表达量上调,绿色代表表达量下调,灰色代表表达量无显著差异,下同
Fig. 8 Expression patterns of BAG family genes in C. sativus L. (A) and C. moschata Duch. (B) under stress HT: High temperature stress. CS: Low temperature stress. Na: Salt stress. The red color indicates the up-regulation of the expression, the green color indicates the down-regulation of the expression, and the gray color indicates the expression is no significant difference in the expression, the same below
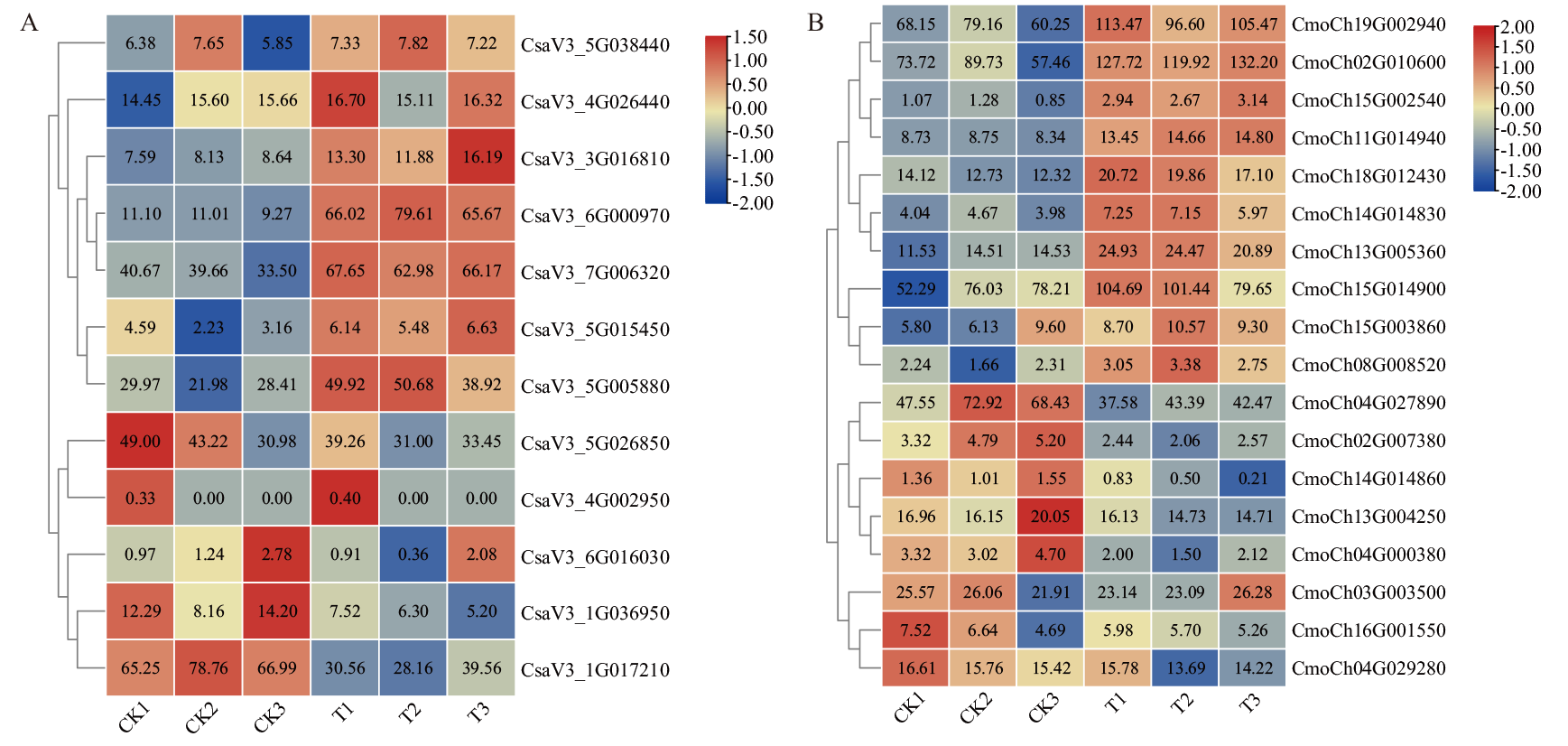
图9 黄瓜(A)和南瓜(B)BAG家族基因在嫁接愈合中的表达热图 CK1、CK2、CK3:黑暗对照组;T1、T2、T3:光照处理组。下同
Fig. 9 Expression heat map of BAG gene family in C. sativus L. (A) and C. moschata Duch. (B) in grafting healing CK1, CK2, CK3: Dark control group. T1, T2, T3: Lighting treatment group. The same below
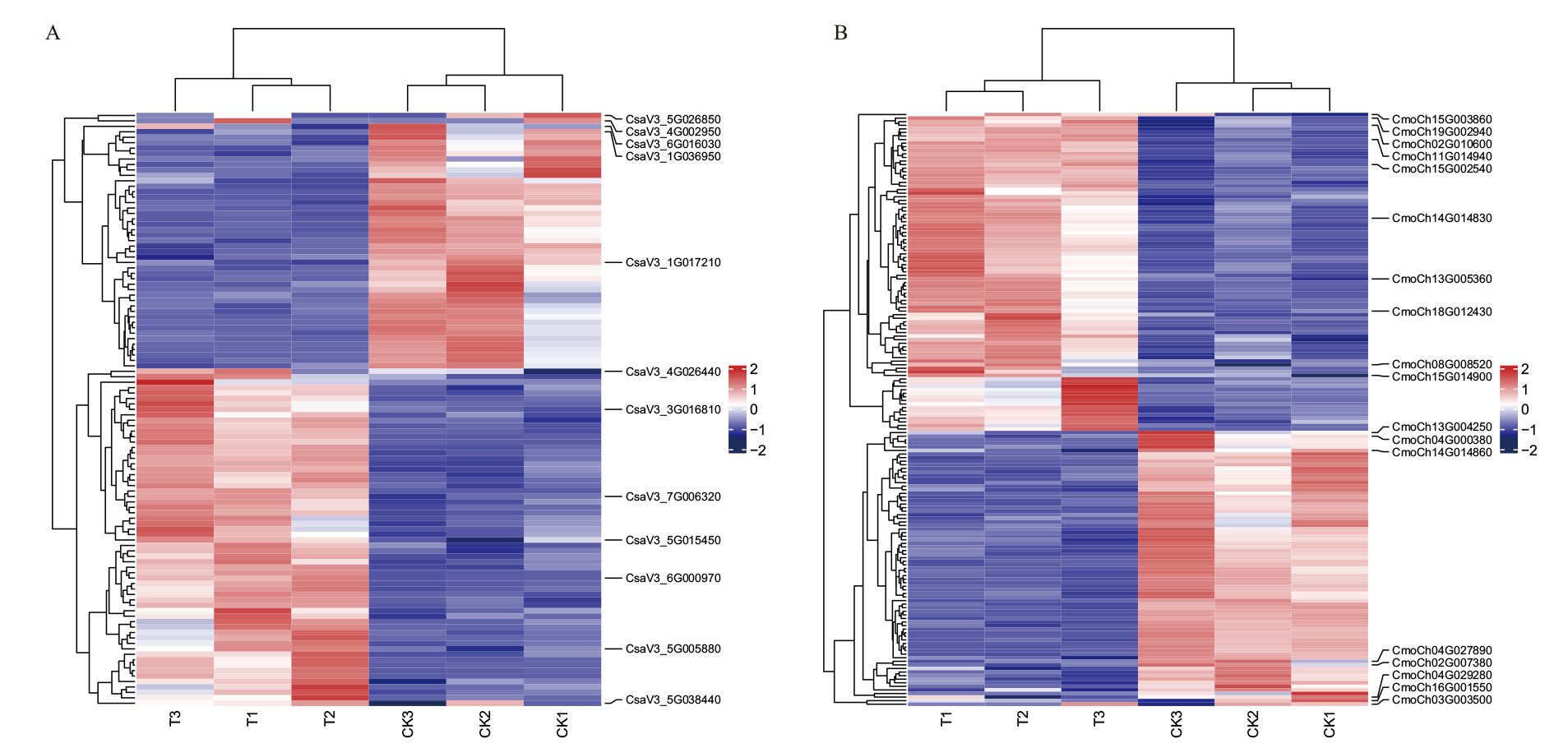
图10 黄瓜(A)和南瓜(B)BAG家族基因与糖代谢通路差异基因的共表达模式热图
Fig. 10 Co-expression patterns of BAG family genes and differential genes in glucose metabolism pathway in C. sativus L. (A) and C. moschata Duch. (B)
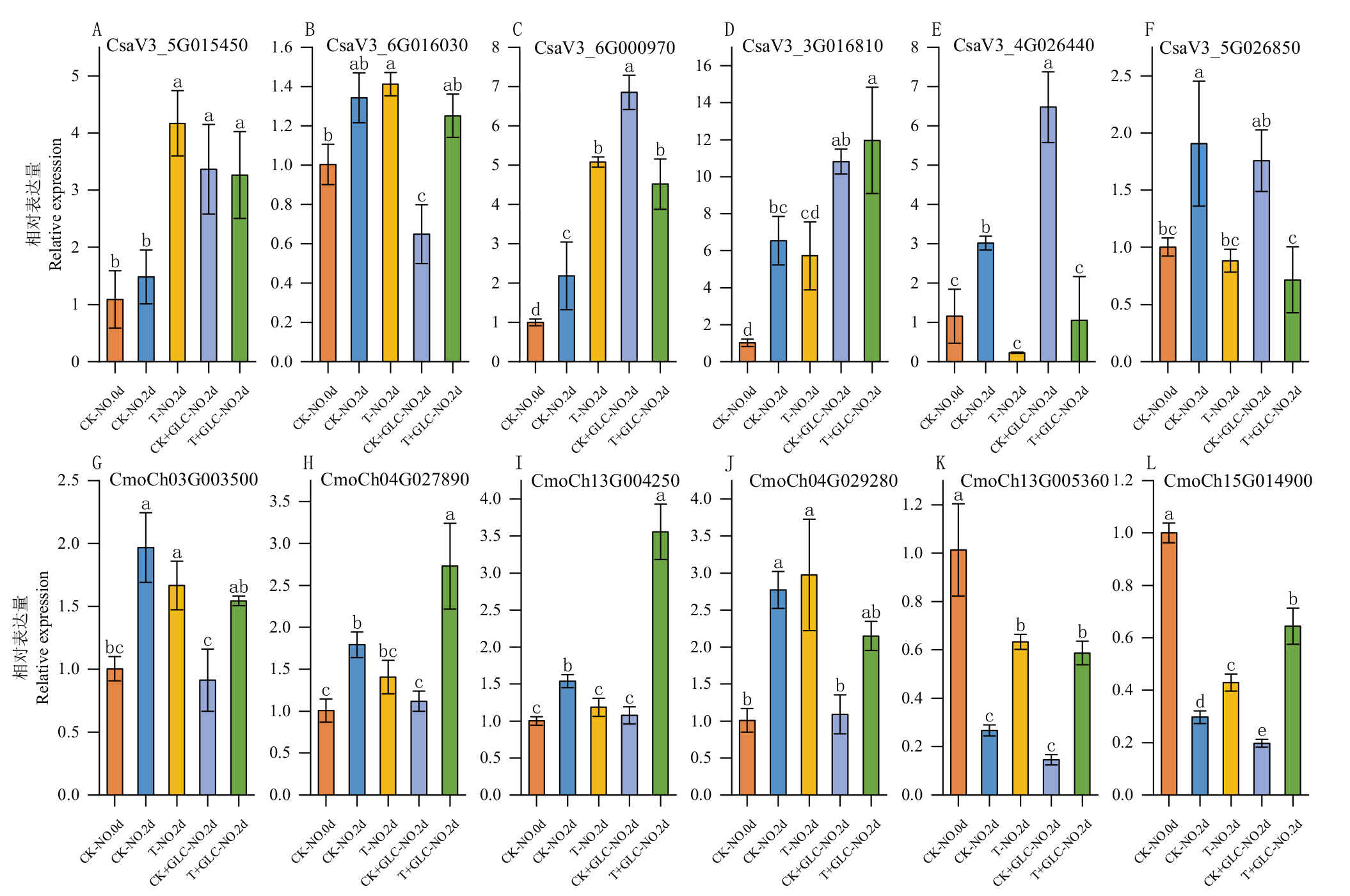
图11 黄瓜(A-F)和南瓜(G-L)BAG家族基因在光照和外源糖处理下的RT-qPCR验证 CK-NO.0d:0 d黑暗对照;CK-NO.2d:2 d黑暗对照;T-NO.2d:2 d光照处理;CK+GLU-NO.2d:2 d黑暗+外源糖;T+GLU-NO.2d:2 d光+外源糖;不同字母表示在P<0.05水平差异显著
Fig. 11 RT-qPCR validation of BAG gene family in C. sativus L. (A-F) and C. moschata Duch. (G-L) under light and exogenous sugar treatments CK-NO.0d: 0 d dark control group. CK-NO. 2d: 2 d dark control group. T-NO.2d: 2 d lighting treatment group. CK+GLU-NO.2d: 2 d dark + sugar treatment group. T+GLU-NO.2d: 2 d light + sugar treatment group. Different letters indicate significant difference at P<0.05 level
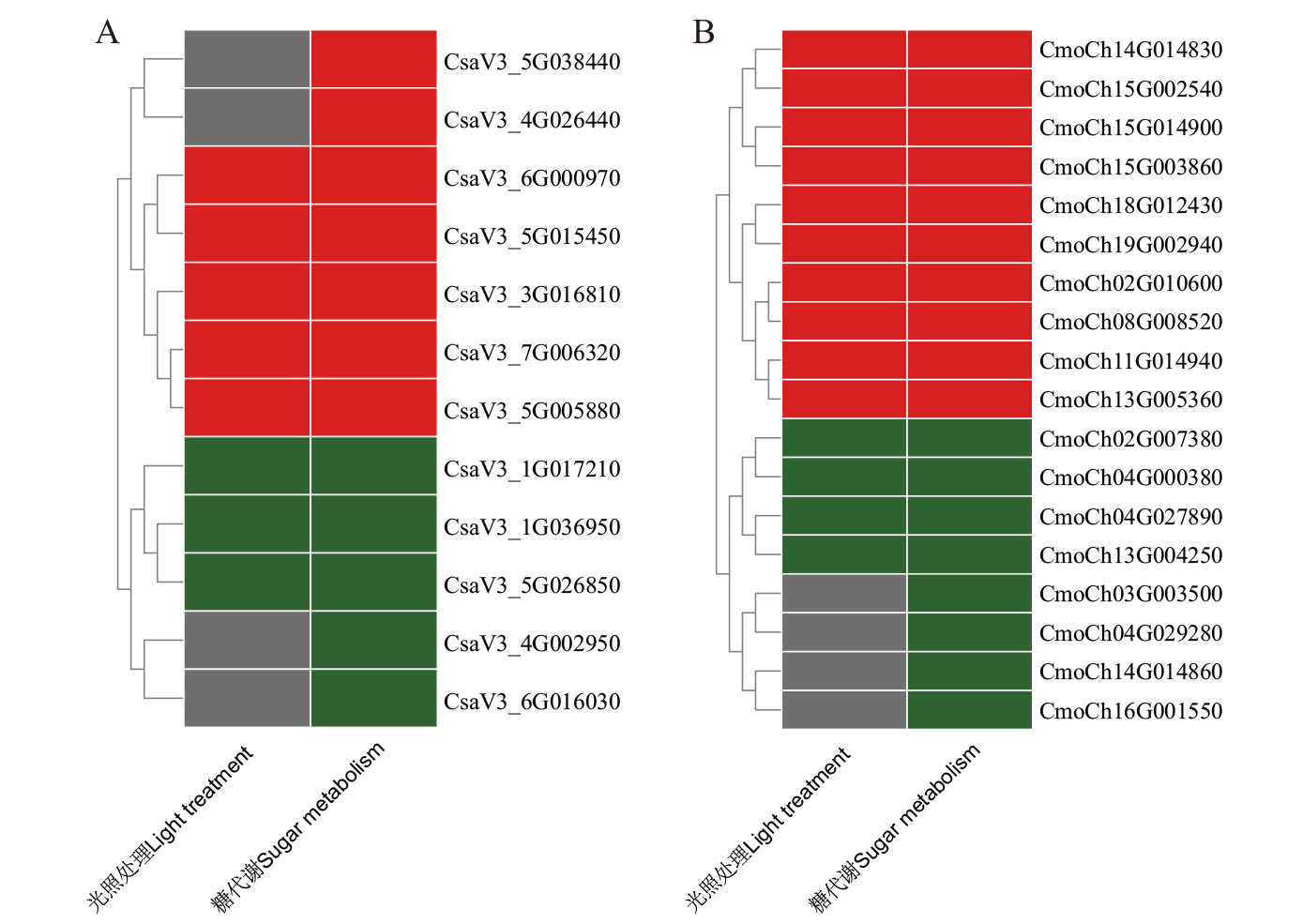
图12 黄瓜(A)和南瓜(B)BAG家族基因在嫁接愈合过程中的表达模式热图 Light:光照处理;Sugar:糖代谢共表达
Fig. 12 Expression patterns of BAG family genes in C. sativus L. (A) and C. moschata Duch. (B) during grafting healing Light: Lighting treatment. Sugar: Co-expression of sugar metabolism
| [1] |
Thanthrige N, J ain S, Bhowmik SD, et al. Centrality of BAGs in plant PCD, stress responses, and host defense[J]. Trends Plant Sci, 2020, 25(11): 1131-1140.
doi: 10.1016/j.tplants.2020.04.012 pmid: 32467063 |
| [2] | Huang SW, Li RQ, Z hang ZH, et al. The genome of the cucumber, Cucumis sativus L.[J]. Nat Genet, 2009, 41(12): 1275-1281. |
| [3] |
Briknarová K, Takayama S, Brive L, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein[J]. Nat Struct Biol, 2001, 8(4): 349-352.
pmid: 11276257 |
| [4] |
Takayama S, Sato T, Krajewski S, et al. Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity[J]. Cell, 1995, 80(2): 279-284.
pmid: 7834747 |
| [5] |
Doukhanina EV, Chen SR, van der Zalm E, et al. Identification and functional characterization of the BAG protein family in Arabidopsis thaliana[J]. J Biol Chem, 2006, 281(27): 18793-18801.
doi: 10.1074/jbc.M511794200 pmid: 16636050 |
| [6] | Poulsen EG, K ampmeyer C, Kriegenburg F, et al. UBL/BAG-domain co-chaperones cause cellular stress upon overexpression through constitutive activation of Hsf1[J]. Cell Stress & Chaperones, 2017, 22(1): 143-154. |
| [7] |
Yang TB, Poovaiah BW. Calcium/calmodulin-mediated signal network in plants[J]. Trends Plant Sci, 2003, 8(10): 505-512.
doi: 10.1016/j.tplants.2003.09.004 pmid: 14557048 |
| [8] | Cui BY, Fang SS, Xing YF, et al. Crystallographic analysis of the Arabidopsis thaliana BAG5-calmodulin protein complex[J]. Acta Crystallogr F Struct Biol Commun, 2015, 71(Pt 7): 870-875. |
| [9] | Chen Y, Wang KK, Di J, et al. Mutation of the BAG-1 domain decreases its protective effect against hypoxia/reoxygenation by regulating HSP70 and the PI3K/AKT signalling pathway in SY-SH5Y cells[J]. Brain Res, 2021, 1751: 147192. |
| [10] | Brenner CM, Choudhary M, McC ormick MG, et al. BAG3: Nature's quintessential multi-functional protein functions as a ubiquitous intra-cellular glue[J]. Cells, 2023, 12(6): 937. |
| [11] | Kang CH, Lee JH, Kim YJ, et al. Characterization of AtBAG2 as a novel molecular chaperone[J]. Life-Basel, 2023, 13(3): 687. |
| [12] | Baeken MW, Behl C. On the origin of BAG(3)and its consequences for an expansion of BAG3's role in protein homeostasis[J]. J Cell Biochem, 2022, 123(1): 102-114. |
| [13] | Gupta MK, Randhawa PK, Masternak MM. Role of BAG 5 in protein quality control: Double-edged sword?[J]. Front Aging, 2022, 3: 844168. |
| [14] | Huang HM, Liu CX, Yang C, et al. BAG9 confers thermotolerance by regulating cellular redox homeostasis and the stability of heat shock proteins in Solanum lycopersicum[J]. Antioxidants, 2022, 11(8): 1467. |
| [15] | 杨美玲, 贺丽霞, 于玮玮, 等. 新疆野苹果BAG家族成员分离及BAG7基因的功能分析[J]. 南开大学学报:自然科学版, 2022, 55(6): 1-6. |
| Yang ML, He LX, Yu WW, et al. Identification of BAG and functional analysis of BAG7 of malus sieversii[J]. Acta Sci Nat Univ Nankaiensis, 2022, 55(6): 1-6. | |
| [16] | Arif M, Li ZT, Luo Q, et al. The BAG2 and BAG6 genes are involved in multiple abiotic stress tolerances in Arabidopsis thaliana[J]. Int J Mol Sci, 2021, 22(11): 5856. |
| [17] | Zhang HH, Li YR, Dickman MB, et al. Cytoprotective co-chaperone BcBAG1 is a component for fungal development, virulence, and unfolded protein response(UPR)of Botrytis cinerea[J]. Front Microbiol, 2019, 10: 685. |
| [18] | Li YR, Williams B, Dickman M. Arabidopsis B-cell lymphoma2(Bcl-2)-associated athanogene 7(BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29[J]. New Phytol, 2017, 214(2): 695-705. |
| [19] |
Li LH, Xing YF, Chang D, et al. CaM/BAG5/Hsc70 signaling complex dynamically regulates leaf senescence[J]. Sci Rep, 2016, 6: 31889.
doi: 10.1038/srep31889 pmid: 27539741 |
| [20] | Li YR, Dickman M. Processing of AtBAG6 triggers autophagy and fungal resistance[J]. Plant Signal Behav, 2016, 11(6): e1175699. |
| [21] | Ding HD, Qian L, Jiang HL, et al. Overexpression of a Bcl-2-associated athanogene SlBAG9 negatively regulates high-temperature response in tomato[J]. Int J Biol Macromol, 2022, 194: 695-705. |
| [22] | Yan JQ, He CX, Zhang H. The BAG-family proteins in Arabidopsis thaliana[J]. Plant Sci, 2003, 165(1): 1-7. |
| [23] | Zhou H, Li JY, Liu XY, et al. The divergent roles of the rice bcl-2 associated athanogene(BAG)genes in plant development and environmental responses[J]. Plants, 2021, 10(10): 2169. |
| [24] |
Ge SM, Kang Z, Li Y, et al. Cloning and function analysis of BAG family genes in wheat[J]. Funct Plant Biol, 2016, 43(5): 393-402.
doi: 10.1071/FP15317 pmid: 32480470 |
| [25] | Yeckel G. Characterization of a soybean BAG gene and its potential role in nematode resistance[D]. Columbia: University of Missouri, 2012. |
| [26] | Jiang HL, Ji YR, Sheng JR, et al. Genome-wide identification of the Bcl-2 associated athanogene(BAG)gene family in Solanum lycopersicum and the functional role of SlBAG9 in response to osmotic stress[J]. Antioxidants, 2022, 11(3): 598. |
| [27] | 莫心怡, 周海连, 何丹丹, 等. 甘蔗割手密BAG基因家族的鉴定与表达分析[J]. 基因组学与应用生物学, 2023, 42(12): 1364-1378. |
| Mo XY, Zhou HL, He DD, et al. Identification and expression analysis of BAG gene family in sugarcane saccharum spontaneum[J]. Genom Appl Biol, 2023, 42(12): 1364-1378. | |
| [28] | 张扬, 杜琳, 唐贤丰, 等. 杨树BAG基因的鉴定及表达模式分析[J]. 林业科学, 2019, 55(1): 138-145. |
| Zhang Y, Du L, Tang XF, et al. Identification and expression pattern analyses of Populus BAG genes[J]. Sci Silvae Sin, 2019, 55(1): 138-145. | |
| [29] | Dash A, Ghag SB. Genome-wide in silico characterization and stress induced expression analysis of BcL-2 associated athanogene(BAG)family in Musa spp[J]. Sci Rep, 2022, 12(1): 625. |
| [30] | Lee DW, Kim SJ, Oh YJ, et al. Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins[J]. Mol Plant, 2016, 9(10): 1428-1431. |
| [31] | You QY, Zhai KR, Yang DL, et al. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance[J]. Cell Host Microbe, 2016, 20(6): 758-769. |
| [32] |
Locascio A, Marqués MC, García-Martínez G, et al. BCL2-associated athanogene4 regulates the KAT1 potassium channel and controls stomatal movement[J]. Plant Physiol, 2019, 181(3): 1277-1294.
doi: 10.1104/pp.19.00224 pmid: 31451552 |
| [33] | He MM, Wang Y, Jahan MS, et al. Characterization of SlBAG genes from Solanum lycopersicum and its function in response to dark-induced leaf senescence[J]. Plants, 2021, 10(5): 947. |
| [34] | Irfan M, Kumar P, Ahmad I, et al. Unraveling the role of tomato Bcl-2-associated athanogene(BAG)proteins during abiotic stress response and fruit ripening[J]. Sci Rep, 2021, 11(1): 21734. |
| [35] | Shang KJ, Xiao L, Zhang XP, et al. Tomato chlorosis virus p22 interacts with NbBAG5 to inhibit autophagy and regulate virus infection[J]. Mol Plant Pathol, 2023, 24(5): 425-435. |
| [36] | Jiang HL, Liu XY, Xiao PX, et al. Functional insights of plant bcl-2-associated ahanogene(BAG)proteins: Multi-taskers in diverse cellular signal transduction pathways[J]. Front Plant Sci, 2023, 14: 1136873. |
| [37] |
张开京, 何帅帅, 贾利, 等. 黄瓜DIR家族基因的全基因组鉴定及其表达分析[J]. 中国农业科学, 2023, 56(4): 711-728.
doi: 10.3864/j.issn.0578-1752.2023.04.010 |
| Zhang KJ, He SS, Jia L, et al. Genome-wide identification and expression analysis of DlR gene family in cucumbers[J]. Sci Agric Sin, 2023, 56(4): 711-728. | |
| [38] | 赵振翔, 敖文红, 王新法, 等. 黄瓜DME基因家族的全基因组鉴别及转录分析[J]. 植物生理学报, 2023, 59(1): 209-218. |
| Zhao ZX, Ao WH, Wang XF, et al. Genome-wide identification and transcriptional analysis of DME gene family in cucumber[J]. Plant Physiol J, 2023, 59(1): 209-218. | |
| [39] | 应奥, 程志华, 张小兰, 等. 黄瓜YABBYs家族及CsYAB1a基因功能初探[J]. 中国农业大学学报, 2022, 27(11): 60-78. |
| Ying A, Cheng ZH, Zhang XL, et al. Preliminary study on the function of YABBYs family and CsYAB1a gene in cucumber.(Cucumis sativus L.)[J]. J China Agric Univ, 2022, 27(11): 60-78. | |
| [40] |
周国彦, 银珊珊, 高佳鑫, 等. 黄瓜AHP基因家族的鉴定及其非生物胁迫表达分析[J]. 生物技术通报, 2022, 38(6): 112-119.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1338 |
| Zhou GY, Yin SS, Gao JX, et al. Identification of AHP gene family in cucumis sativus and its expression analysis under abiotic stress[J]. Biotechnol Bull, 2022, 38(6): 112-119. | |
| [41] | Xu MY, Wang YP, Zhang MT, et al. Genome-wide identification of BES1 gene family in six cucurbitaceae species and its expression analysis in Cucurbita moschata[J]. Int J Mol Sci, 2023, 24(3): 2287. |
| [42] | Davoudi M, Chen JF, Lou QF. Genome-wide identification and expression analysis of heat shock protein 70(HSP70)gene family in pumpkin(Cucurbita moschata)rootstock under drought stress suggested the potential role of these chaperones in stress tolerance[J]. Int J Mol Sci, 2022, 23(3): 1918. |
| [43] | Hu YP, Zhang TT, Liu Y, et al. Pumpkin(Cucurbita moschata)HSP20 gene family identification and expression under heat stress[J]. Front Genet, 2021, 12: 753953. |
| [44] | Miao L, Li SZ, Shi AK, et al. Genome-wide analysis of the AINTEGUMENTA-like(AIL)transcription factor gene family in pumpkin(Cucurbita moschata Duch.) and CmoANT1.2 response in graft union healing[J]. Plant Physiol Biochem, 2021, 162: 706-715. |
| [45] | Li Q, Li HB, Huang W, et al. A chromosome-scale genome assembly of cucumber(Cucumis sativus L.)[J]. Gigascience, 2019, 8(6): giz072. |
| [46] |
Sun HH, Wu S, Zhang GY, et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid cucurbita genomes[J]. Mol Plant, 2017, 10(10): 1293-1306.
doi: S1674-2052(17)30266-6 pmid: 28917590 |
| [47] | Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching[J]. Nucleic Acids Res, 2011, 39(Web Server issue): W29-W37. |
| [48] | Mistry J, Chuguransky S, Williams L, et al. Pfam: The protein families database in 2021[J]. Nucleic Acids Res, 2021, 49(D1): D412-D419. |
| [49] | Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020[J]. Nucleic Acids Res, 2021, 49(D1): D458-D460. |
| [50] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [51] | Bailey TL, Williams N, Misleh C, et al. MEME: discovering and analyzing DNA and protein sequence motifs[J]. Nucleic Acids Res, 2006, 34(Web Server issue): W369-W373. |
| [52] |
Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11[J]. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [53] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [54] | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity[J]. Nucleic Acids Res, 2012, 40(7): e49. |
| [55] |
Li Z, Zhang ZH, Yan PC, et al. RNA-Seq improves annotation of protein-coding genes in the cucumber genome[J]. BMC Genomics, 2011, 12: 540.
doi: 10.1186/1471-2164-12-540 pmid: 22047402 |
| [56] | Chen CH, Chen XQ, Han J, et al. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses[J]. BMC Plant Biol, 2020, 20(1): 443. |
| [57] | Zhu Y, Yin JL, Liang YF, et al. Transcriptomic dynamics provide an insight into the mechanism for silicon-mediated alleviation of salt stress in cucumber plants[J]. Ecotoxicol Environ Saf, 2019, 174: 245-254. |
| [58] |
Zhou Y, Yang K, Cheng M, et al. Double-faced role of Bcl-2-associated athanogene 7 in plant-Phytophthora interaction[J]. J Exp Bot, 2021, 72(15): 5751-5765.
doi: 10.1093/jxb/erab252 pmid: 34195821 |
| [59] | Hu LZ, Chen JT, Guo JJ, et al. Functional divergence and evolutionary dynamics of BAG gene family in maize(Zea mays)[J]. Int J Agric Biol, 2013, 15(2): 200-206. |
| [60] | UN Food and Agriculture Organization, Corporate Statistical Database(FAOSTAT). Production of cucumbers and gherkins in 2020[EB/OL]. https://www.fao.org/faostat/zh/#data/.2020-03-30/2023-12-18. |
| [61] | Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice(Oryza sativa)[J]. Genomics, 2006, 88(3): 360-371. |
| [62] |
Zhang SB, C hen C, Li L, et al. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family[J]. Plant Physiol, 2005, 139(3): 1107-1124.
pmid: 16286450 |
| [63] |
Nollen EA, Brunsting JF, Song J, et al. Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity[J]. Mol Cell Biol, 2000, 20(3): 1083-1088.
doi: 10.1128/MCB.20.3.1083-1088.2000 pmid: 10629065 |
| [64] |
Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators[J]. J Biol Chem, 1999, 274(2): 781-786.
doi: 10.1074/jbc.274.2.781 pmid: 9873016 |
| [65] |
Takayama S, Bimston DN, Matsuzawa S, et al. BAG-1 modulates the chaperone activity of Hsp70/Hsc70[J]. EMBO J, 1997, 16(16): 4887-4896.
doi: 10.1093/emboj/16.16.4887 pmid: 9305631 |
| [66] | Nawkar GM, Maibam P, Park JH, et al. In silico study on Arabidopsis BAG gene expression in response to environmental stresses[J]. Protoplasma, 2017, 254(1): 409-421. |
| [67] | Wang J, Nan N, Li N, et al. A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress[J]. Plant Cell, 2020, 32(11): 3535-3558. |
| [68] |
Wang JY, Yeckel G, Kandoth PK, et al. Targeted suppression of soybean BAG6-induced cell death in yeast by soybean cyst nematode effectors[J]. Mol Plant Pathol, 2020, 21(9): 1227-1239.
doi: 10.1111/mpp.12970 pmid: 32686295 |
| [1] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [2] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [3] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [4] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [5] | 侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178. |
| [6] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [7] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [8] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [9] | 张清兰, 张亚冉, 鞠志花, 王秀革, 肖遥, 王金鹏, 魏晓超, 高亚平, 白福恒, 王洪程. 牛TARDBP基因核心启动子鉴定与转录调控分析[J]. 生物技术通报, 2024, 40(4): 306-318. |
| [10] | 陈晓松, 刘超杰, 郑佳, 乔宗伟, 罗惠波, 邹伟. TMT定量蛋白质组学解析Rummeliibacillus suwonensis 3B-1 生长及己酸代谢机制[J]. 生物技术通报, 2024, 40(3): 135-145. |
| [11] | 杨伟杰, 杨周林, 朱浩东, 魏煜, 刘君, 刘训. 地衣素合成酶关键模块 LchAD 蛋白的性质和功能研究[J]. 生物技术通报, 2024, 40(3): 322-332. |
| [12] | 龚丽丽, 余花, 杨杰, 陈天池, 赵双滢, 吴月燕. 葡萄CYP707A基因家族的鉴定及对果实成熟的功能验证[J]. 生物技术通报, 2024, 40(2): 160-171. |
| [13] | 褚睿, 李昭轩, 张学青, 杨东亚, 曹行行, 张雪艳. 黄瓜枯萎病拮抗芽孢杆菌的筛选、鉴定及其生防潜力[J]. 生物技术通报, 2023, 39(8): 262-271. |
| [14] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [15] | 李敬蕊, 王育博, 解紫薇, 李畅, 吴晓蕾, 宫彬彬, 高洪波. 甜瓜PIN基因家族的鉴定及高温胁迫表达分析[J]. 生物技术通报, 2023, 39(5): 192-204. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||