








生物技术通报 ›› 2025, Vol. 41 ›› Issue (2): 257-269.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0700
• 研究报告 • 上一篇
杨涌( ), 袁国梅(
), 袁国梅( ), 康肖肖, 刘亚明, 王东升, 张海娥(
), 康肖肖, 刘亚明, 王东升, 张海娥( )
)
收稿日期:2024-07-21
出版日期:2025-02-26
发布日期:2025-02-28
通讯作者:
张海娥,女,博士,研究员,研究方向 :板栗育种;E-mail: zhang33haie4@163.com作者简介:杨涌,男,硕士研究生,研究方向 :板栗育种;E-mail: 1725722566@qq.com基金资助:
YANG Yong( ), YUAN Guo-mei(
), YUAN Guo-mei( ), KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e(
), KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e( )
)
Received:2024-07-21
Published:2025-02-26
Online:2025-02-28
摘要:
目的 SWEET基因家族与果实中淀粉含量、糖含量密切相关,分析板栗SWEET基因家族在板栗果实成熟过程中的表达模式,为后续研究板栗SWEET基因家族在果实成熟过程中的作用奠定基础。 方法 利用生物信息学方法鉴定出7种壳斗科物种SWEET基因家族,对其进行系统进化树构建、各个成员同源性分析、基因复制类型分析等;同时分析板栗SWEET基因家族蛋白理化性质、基因结构、选择压力、蛋白结构等;利用转录组和RT-qPCR分析板栗SWEET基因家族成员在不同果实成熟时期的表达模式。 结果 7种壳斗科物种中共包含了129个SWEET基因家族成员,7种壳斗科物种分化之前共拥有16个SWEET基因家族成员。板栗SWEET基因家族共有19个,不均地分布在9条染色体和1条contig片段上;同时其蛋白拥有较为一致的Motif分布、保守结构域分布。顺势作用元件分析结果显示,大量的生长发育相关元件、激素响应元件以及胁迫响应元件存在于板栗SWEET基因家族启动子序列上。转录组和RT-qPCR分析显示,有9个板栗SWEET基因家族成员在板栗果实成熟过程中表达。其中CmSWEET1、CmSWEET3、CmSWEET4、CmSWEET16及CmSWEET19随着果实成熟过程基因表达量下调;CmSWEET2及CmSWEET9随着果实成熟过程基因表达量上调;CmSWEET15与CmSWEET17随着果实成熟过程基因表达量先下调再上调。 结论 共鉴定了129个壳斗科物种SWEET基因家族成员,其中有19个板栗SWEET基因家族成员,CmSWEET15与CmSWEET17表达模式与可溶性糖变化模式相似,其可能调控板栗果实成熟过程中可溶性糖转运;CmSWEET1、CmSWEET2、CmSWEET3、CmSWEET4、CmSWEET9、CmSWEET16及CmSWEET19表达模式与淀粉变化模式完全一致或完全相反,其可能调控板栗果实成熟过程中淀粉积累。
杨涌, 袁国梅, 康肖肖, 刘亚明, 王东升, 张海娥. 板栗SWEET基因家族成员的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 257-269.
YANG Yong, YUAN Guo-mei, KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e. Identification and Expression Analysis of Members of the SWEET Gene Family in Chinese Chestnut[J]. Biotechnology Bulletin, 2025, 41(2): 257-269.
| 基因Gene | 上游引物Forward primer(5′-3′) | 下游引物Reverse primer(5′-3′) |
|---|---|---|
| CmSWEET1 | CTGCATGGTGTGGGCCTT | GATACCAGTGCCTGCCCC |
| CmSWEET2 | GCGCTGTTTGTGTCACCC | AGCAGCTGAAGAAGGCGT |
| CmSWEET3 | CAAAGGAAGCAGCCTCCCA | TGGGGCCAACTAGGGTGT |
| CmSWEET9 | ATGCAGCTGGAGTCGCAG | CGGCAGCCCTGAGAACTG |
| CmSWEET14 | GCTTTGCTGCTGCCGTTT | CAGGAAGTGCCGCACAGA |
| CmSWEET15 | TCGCGTTTGTGCTCACCT | AAACGGCAGCAGCAAAGC |
| CmSWEET16 | CAGCCGGGAGAGCTAGGA | AGCAGGGAGGTGCAAAGC |
| CmSWEET17 | GCACCCGGGAGAGTTAGG | AGCAGGGAGGTGCAAAGC |
| CmSWEET19 | CCTGGCAAAAGGCTCCCA | TGGCATGTACTCCACGCTC |
| CmActin | ATTCACGAGACCACCTACA | TGCCACAACCTTAATCTTCAT |
表1 引物序列
Table 1 Primer sequences
| 基因Gene | 上游引物Forward primer(5′-3′) | 下游引物Reverse primer(5′-3′) |
|---|---|---|
| CmSWEET1 | CTGCATGGTGTGGGCCTT | GATACCAGTGCCTGCCCC |
| CmSWEET2 | GCGCTGTTTGTGTCACCC | AGCAGCTGAAGAAGGCGT |
| CmSWEET3 | CAAAGGAAGCAGCCTCCCA | TGGGGCCAACTAGGGTGT |
| CmSWEET9 | ATGCAGCTGGAGTCGCAG | CGGCAGCCCTGAGAACTG |
| CmSWEET14 | GCTTTGCTGCTGCCGTTT | CAGGAAGTGCCGCACAGA |
| CmSWEET15 | TCGCGTTTGTGCTCACCT | AAACGGCAGCAGCAAAGC |
| CmSWEET16 | CAGCCGGGAGAGCTAGGA | AGCAGGGAGGTGCAAAGC |
| CmSWEET17 | GCACCCGGGAGAGTTAGG | AGCAGGGAGGTGCAAAGC |
| CmSWEET19 | CCTGGCAAAAGGCTCCCA | TGGCATGTACTCCACGCTC |
| CmActin | ATTCACGAGACCACCTACA | TGCCACAACCTTAATCTTCAT |

图1 12个物种SWEET基因家族系统进化树图中不同分支颜色代表不同分组;基因名称的不同颜色代表不同同源性分组
Fig. 1 Phylogenetic tree of SWEET gene family of 12 speciesDifferent branch colors in the figure indicate different groups; different colors of gene names indicate different orthogroups
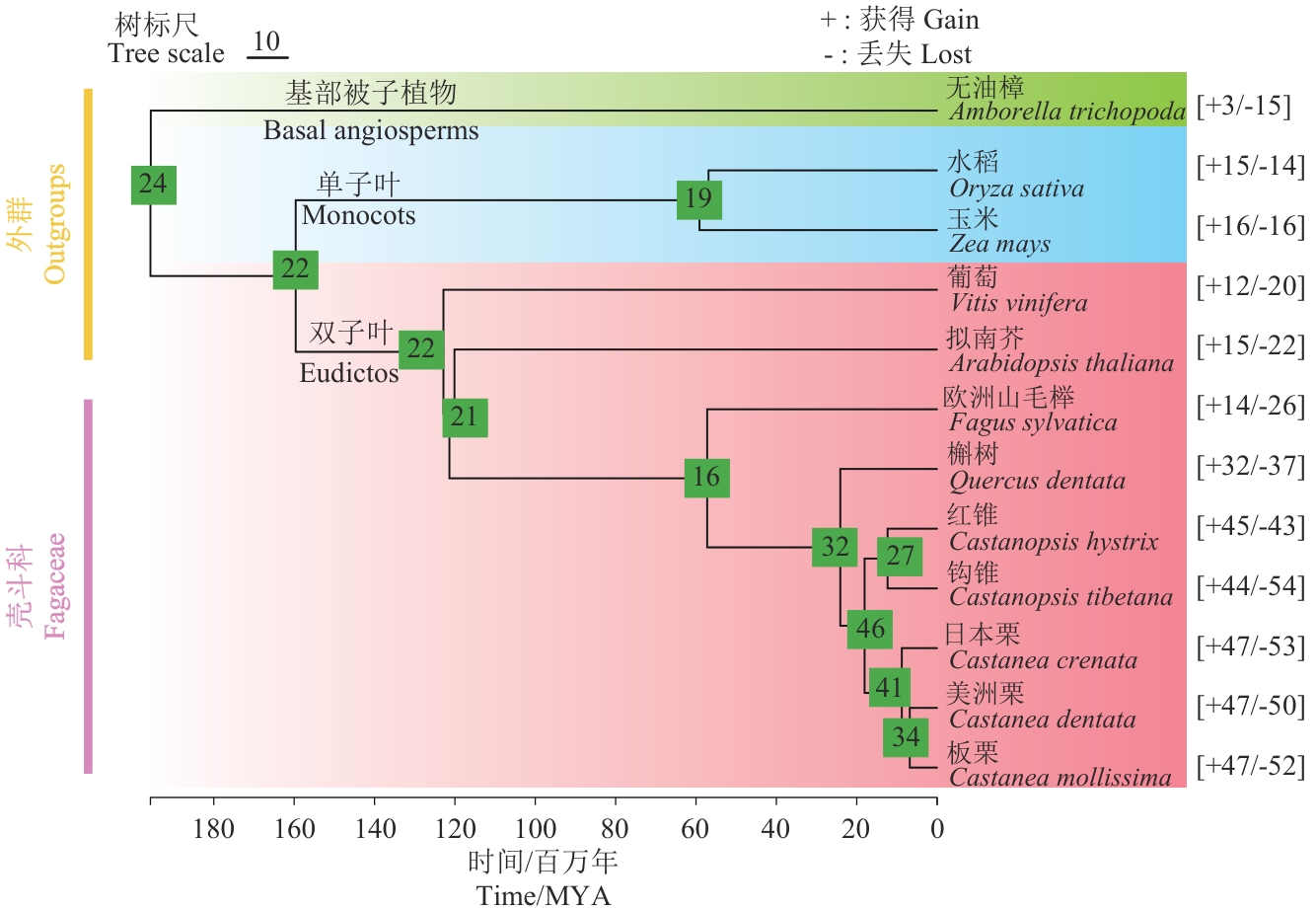
图3 各进化节点基因家族成员数量统计图中均为绿色方框代表各个节点SWEET基因家族成员数量;“+,-”后数字代表获得丢失的基因数量
Fig. 3 Statistics of the number of gene family members at each evolutionary nodeThe green boxes indicate the number of members of the SWEET gene family in each node. Numbers after “+, -” indicate the number of gained or lost genes
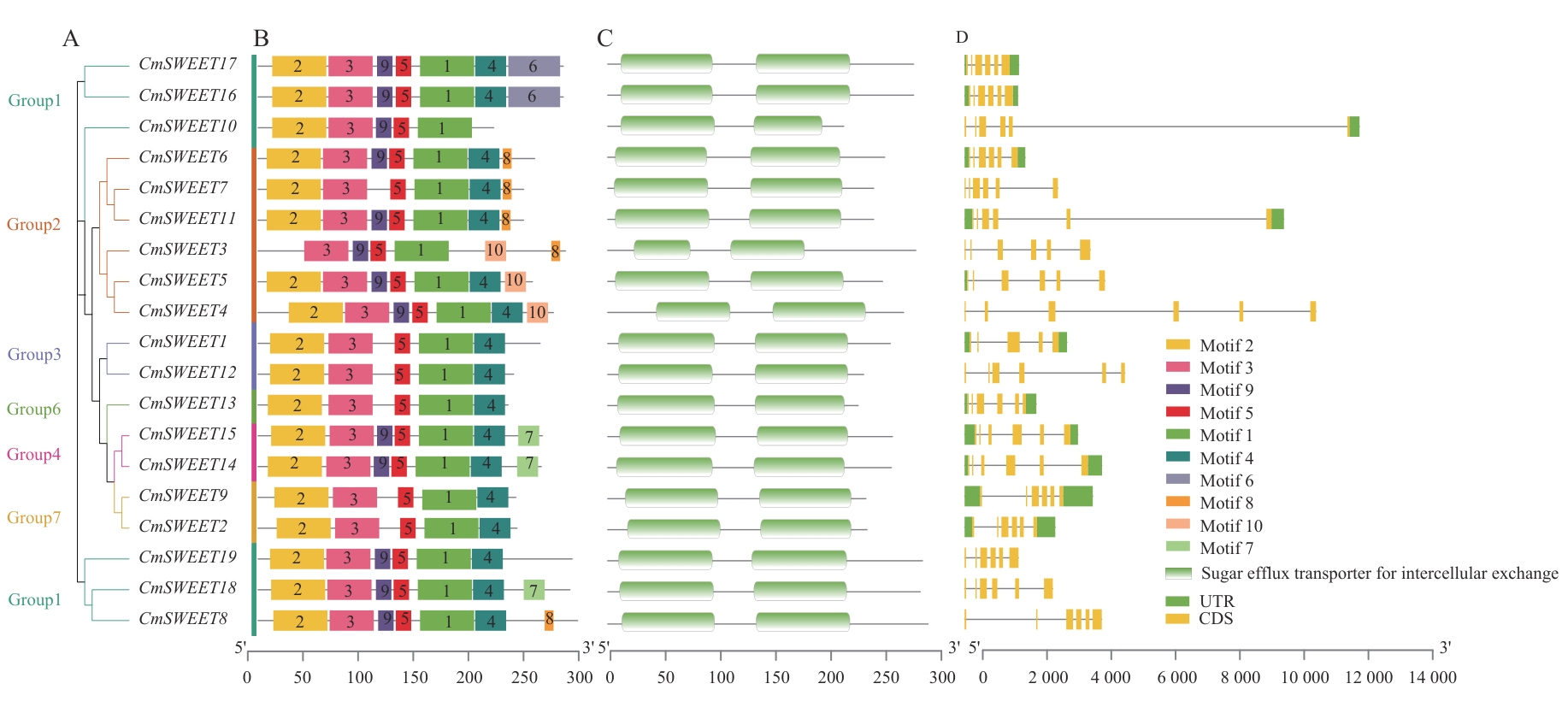
图5 CmSWEET基因家族系统进化树(A)、motif分布(B)、结构域分布(C)和基因结构(D)
Fig. 5 Phylogenic tree (A), motif distribution (B), domain distribution (C), and gene structure (D) of CmSWEET gene family
| 物种 Species | 全基因组复制 WGD | 串联复制 TD | 近端复制 PD | 分散复制 DSD | 转座子复制 TRD | 总数 Total |
|---|---|---|---|---|---|---|
| 欧洲山毛榉 Fagus sylvatica | 1 | 0 | 2 | 1 | 2 | 6 |
| 槲树 Quercus dentata | 3 | 2 | 1 | 1 | 3 | 10 |
| 红锥 Castanopsis hystrix | 2 | 4 | 3 | 0 | 5 | 14 |
| 钩椎 Castanopsis tibetana | 1 | 1 | 2 | 0 | 4 | 8 |
| 日本栗 Castanea crenata | 1 | 1 | 2 | 3 | 4 | 11 |
| 美洲栗 Castanea dentata | 3 | 5 | 1 | 0 | 2 | 11 |
| 板栗 Castanea mollissima | 0 | 5 | 1 | 3 | 3 | 12 |
表2 七种壳斗科物种SWEET基因家族复制类型
Table 2 SWEET gene family duplication types in seven Fagaceae species
| 物种 Species | 全基因组复制 WGD | 串联复制 TD | 近端复制 PD | 分散复制 DSD | 转座子复制 TRD | 总数 Total |
|---|---|---|---|---|---|---|
| 欧洲山毛榉 Fagus sylvatica | 1 | 0 | 2 | 1 | 2 | 6 |
| 槲树 Quercus dentata | 3 | 2 | 1 | 1 | 3 | 10 |
| 红锥 Castanopsis hystrix | 2 | 4 | 3 | 0 | 5 | 14 |
| 钩椎 Castanopsis tibetana | 1 | 1 | 2 | 0 | 4 | 8 |
| 日本栗 Castanea crenata | 1 | 1 | 2 | 3 | 4 | 11 |
| 美洲栗 Castanea dentata | 3 | 5 | 1 | 0 | 2 | 11 |
| 板栗 Castanea mollissima | 0 | 5 | 1 | 3 | 3 | 12 |
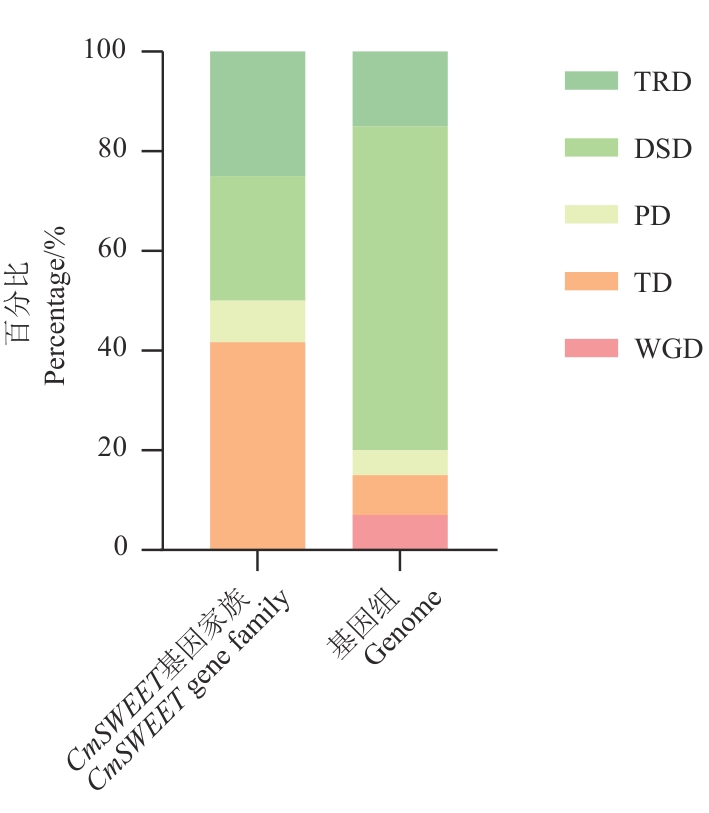
图7 CmSWEET基因家族与基因组复制模式占比比较图中TRD、DSD、PD、TD及WGD代表转座子复制、分散复制、近端复制、串联复制及全基因组复制。下同
Fig. 7 Comparison of the proportion of CmSWEET gene family and genome replication typesTRD, DSD, PD, TD and WGD indicate transposon duplication, dispersed duplication, proximal duplication, tandem duplication and whole genome duplication in the figure. The same below
| 复制类型Duplication type | 基因对Gene pair | 非同义替换率Ka | 同义替换率Ks | 非同义替换率/同义替换率Ka/Ks | |
|---|---|---|---|---|---|
| TD | CmSWEET5 | CmSWEET6 | 0.44 | 4.81 | 0.09 |
| TD | CmSWEET18 | CmSWEET19 | 0.34 | 1.77 | 0.19 |
| TD | CmSWEET6 | CmSWEET7 | 0.37 | 1.07 | 0.34 |
| TD | CmSWEET14 | CmSWEET15 | 0.04 | 0.12 | 0.37 |
| TD | CmSWEET4 | CmSWEET5 | 0.22 | 0.45 | 0.49 |
| PD | CmSWEET16 | CmSWEET17 | 0.02 | 0.06 | 0.28 |
| DSD | CmSWEET8 | CmSWEET11 | 0.64 | 1.69 | 0.38 |
| DSD | CmSWEET13 | CmSWEET11 | 0.57 | 3.47 | 0.16 |
| DSD | CmSWEET11 | CmSWEET3 | 0.45 | 2.14 | 0.21 |
| TRD | CmSWEET9 | CmSWEET2 | 0.23 | 0.98 | 0.23 |
| TRD | CmSWEET1 | CmSWEET12 | 0.42 | 3.06 | 0.14 |
| TRD | CmSWEET10 | CmSWEET17 | 0.33 | 1.73 | 0.19 |
表3 基因对Ka/Ks值的计算
Table 3 Calculations of the gene-pairs Ka/Ks values
| 复制类型Duplication type | 基因对Gene pair | 非同义替换率Ka | 同义替换率Ks | 非同义替换率/同义替换率Ka/Ks | |
|---|---|---|---|---|---|
| TD | CmSWEET5 | CmSWEET6 | 0.44 | 4.81 | 0.09 |
| TD | CmSWEET18 | CmSWEET19 | 0.34 | 1.77 | 0.19 |
| TD | CmSWEET6 | CmSWEET7 | 0.37 | 1.07 | 0.34 |
| TD | CmSWEET14 | CmSWEET15 | 0.04 | 0.12 | 0.37 |
| TD | CmSWEET4 | CmSWEET5 | 0.22 | 0.45 | 0.49 |
| PD | CmSWEET16 | CmSWEET17 | 0.02 | 0.06 | 0.28 |
| DSD | CmSWEET8 | CmSWEET11 | 0.64 | 1.69 | 0.38 |
| DSD | CmSWEET13 | CmSWEET11 | 0.57 | 3.47 | 0.16 |
| DSD | CmSWEET11 | CmSWEET3 | 0.45 | 2.14 | 0.21 |
| TRD | CmSWEET9 | CmSWEET2 | 0.23 | 0.98 | 0.23 |
| TRD | CmSWEET1 | CmSWEET12 | 0.42 | 3.06 | 0.14 |
| TRD | CmSWEET10 | CmSWEET17 | 0.33 | 1.73 | 0.19 |
| 蛋白编号Protein ID | 预测模型得分pTM scores |
|---|---|
| CmSWEET1 | 0.84 |
| CmSWEET2 | 0.86 |
| CmSWEET3 | 0.62 |
| CmSWEET4 | 0.75 |
| CmSWEET5 | 0.81 |
| CmSWEET6 | 0.77 |
| CmSWEET7 | 0.75 |
| CmSWEET8 | 0.69 |
| CmSWEET9 | 0.87 |
| CmSWEET10 | 0.70 |
| CmSWEET11 | 0.81 |
| CmSWEET12 | 0.87 |
| CmSWEET13 | 0.85 |
| CmSWEET14 | 0.79 |
| CmSWEET15 | 0.79 |
| CmSWEET16 | 0.73 |
| CmSWEET17 | 0.72 |
| CmSWEET18 | 0.70 |
| CmSWEET19 | 0.72 |
表4 CmSWEET基因家族蛋白预测模型得分
Table 4 Prediction model scores of CmSWEET gene family proteins
| 蛋白编号Protein ID | 预测模型得分pTM scores |
|---|---|
| CmSWEET1 | 0.84 |
| CmSWEET2 | 0.86 |
| CmSWEET3 | 0.62 |
| CmSWEET4 | 0.75 |
| CmSWEET5 | 0.81 |
| CmSWEET6 | 0.77 |
| CmSWEET7 | 0.75 |
| CmSWEET8 | 0.69 |
| CmSWEET9 | 0.87 |
| CmSWEET10 | 0.70 |
| CmSWEET11 | 0.81 |
| CmSWEET12 | 0.87 |
| CmSWEET13 | 0.85 |
| CmSWEET14 | 0.79 |
| CmSWEET15 | 0.79 |
| CmSWEET16 | 0.73 |
| CmSWEET17 | 0.72 |
| CmSWEET18 | 0.70 |
| CmSWEET19 | 0.72 |
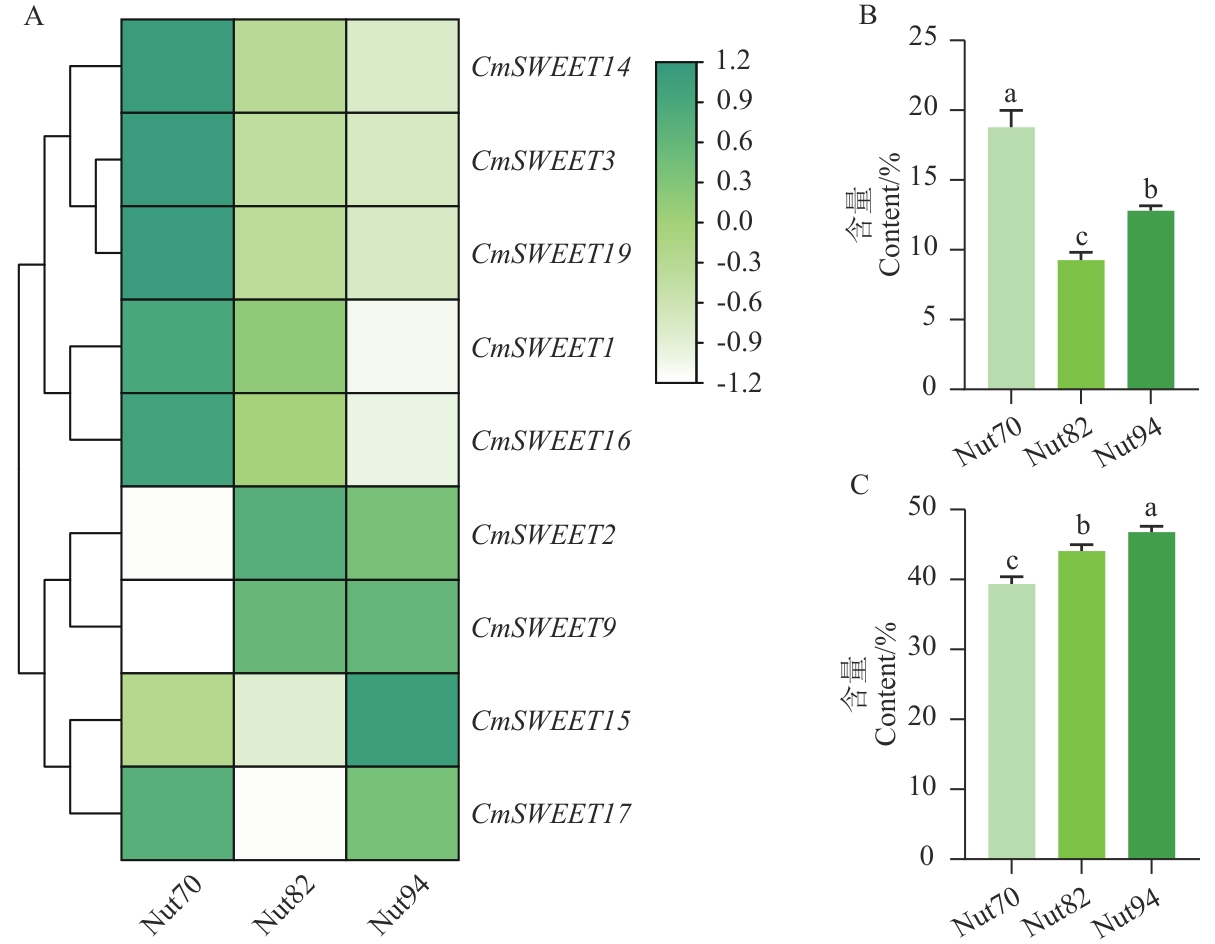
图9 CmSWEET基因家族表达模式热图(A)、可溶性糖含量(B)和淀粉含量(C)变化趋势Nut70代表果实成熟初时期;Nut82代表果实成熟中期;Nut94代表果实成熟后期;不同小写字母表示在0.05水平差异显著;下同
Fig. 9 Heatmap of the CmSWEET gene family expression patterns (A), the changing trend of soluble sugar content (B) and the starch content (C)Nut 70 indicates the early ripening period; Nut 82 indicates the middle ripening stage and Nut 94 indicates the late ripening stage. Different lower letters indicate significantly difference at the 0.05 level. The same below
| 1 | Yamada K, Osakabe Y. Sugar compartmentation as an environmental stress adaptation strategy in plants [J]. Semin Cell Dev Biol, 2018, 83: 106-114. |
| 2 | Pommerrenig B, Ludewig F, Cvetkovic J, et al. In concert: orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance [J]. Plant Cell Physiol, 2018, 59(7): 1290-1299. |
| 3 | Chen LQ, Cheung LS, Feng L, et al. Transport of sugars [J]. Annu Rev Biochem, 2015, 84: 865-894. |
| 4 | Eom JS, Chen LQ, Sosso D, et al. SWEETs, transporters for intracellular and intercellular sugar translocation [J]. Curr Opin Plant Biol, 2015, 25: 53-62. |
| 5 | Xie HH, Wang D, Qin YQ, et al. Genome-wide identification and expression analysis of SWEET gene family in Litchi chinensis reveal the involvement of LcSWEET2a/3b in early seed development [J]. BMC Plant Biol, 2019, 19(1): 499. |
| 6 | Chen LQ, Qu XQ, Hou BH, et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport [J]. Science, 2012, 335(6065): 207-211. |
| 7 | Zhu JL, Zhou L, Li TF, et al. Genome-wide investigation and characterization of SWEET gene family with focus on their evolution and expression during hormone and abiotic stress response in maize [J]. Genes, 2022, 13(10): 1682. |
| 8 | Li M, Xie HJ, He MM, et al. Genome-wide identification and expression analysis of the StSWEET family genes in potato (Solanum tuberosum L.) [J]. Genes Genomics, 2020, 42(2): 135-153. |
| 9 | Hao L, Shi X, Qin SW, et al. Genome-wide identification, characterization and transcriptional profile of the SWEET gene family in Dendrobium officinale [J]. BMC Genomics, 2023, 24(1): 378. |
| 10 | Han XW, Han S, Zhu YX, et al. Genome-wide identification and expression analysis of the SWEET gene family in Capsicum annuum L [J]. Int J Mol Sci, 2023, 24(24): 17408. |
| 11 | Zhang XH, Wang S, Ren Y, et al. Identification, analysis and gene cloning of the SWEET gene family provide insights into sugar transport in pomegranate (Punica granatum) [J]. Int J Mol Sci, 2022, 23(5): 2471. |
| 12 | Li P, Wang LH, Liu HB, et al. Impaired SWEET-mediated sugar transportation impacts starch metabolism in developing rice seeds [J]. Crop J, 2022, 10(1): 98-108. |
| 13 | Radchuk V, Belew ZM, Gündel A, et al. SWEET11b transports both sugar and cytokinin in developing barley grains [J]. Plant Cell, 2023, 35(6): 2186-2207. |
| 14 | Zhang XS, Feng CY, Wang MN, et al. Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits [J]. Hortic Res, 2021, 8(1): 186. |
| 15 | Wang SD, Yokosho K, Guo RZ, et al. The soybean sugar transporter GmSWEET15 mediates sucrose export from endosperm to early embryo [J]. Plant Physiol, 2019, 180(4): 2133-2141. |
| 16 | Zhang SH, Wang H, Wang T, et al. Abscisic acid and regulation of the sugar transporter gene MdSWEET9b promote apple sugar accumulation [J]. Plant Physiol, 2023, 192(3): 2081-2101. |
| 17 | Massantini R, Moscetti R, Frangipane MT. Evaluating progress of chestnut quality: a review of recent developments [J]. Trends Food Sci Technol, 2021, 113: 245-254. |
| 18 | Warmund MR. Chinese chestnut (Castanea mollissima) as a niche crop in the central region of the United States [J]. HortScience, 2011, 46(3): 345-347. |
| 19 | Haytowitz D, Ahuja J, Wu X, et al. USDA National Nutrient Database for standard reference, legacy [EB/OL] . . |
| 20 | Huang RM, Peng F, Wang DS, et al. Transcriptome analysis of differential sugar accumulation in the developing embryo of contrasting two Castanea mollissima cultivars [J]. Front Plant Sci, 2023, 14: 1206585. |
| 21 | Rozewicki J, Li SL, Amada KM, et al. MAFFT-DASH: integrated protein sequence and structural alignment [J]. Nucleic Acids Res, 2019, 47(W1): W5-W10. |
| 22 | Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments [J]. PLoS One, 2010, 5(3): e9490. |
| 23 | Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics [J]. Genome Biol, 2019, 20(1): 238. |
| 24 | Xie JM, Chen YR, Cai GJ, et al. Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees [J]. Nucleic Acids Res, 2023, 51(W1): W587-W592. |
| 25 | Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees [J]. J Comput Biol, 2000, 7(3/4): 429-447. |
| 26 | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| 27 | Wang YP, Tang HB, Debarry JD, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity [J]. Nucleic Acids Res, 2012, 40(7): e49. |
| 28 | Qiao X, Li QH, Yin H, et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants [J]. Genome Biol, 2019, 20(1): 38. |
| 29 | Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3 [J]. Nature, 2024, 630(8016): 493-500. |
| 30 | Li SX, Shi ZG, Zhu QR, et al. Transcriptome sequencing and differential expression analysis of seed starch accumulation in Chinese chestnut Metaxenia [J]. BMC Genomics, 2021, 22(1): 617. |
| 31 | Singh J, Das S, Jagadis Gupta K, et al. Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield [J]. Plant Biotechnol J, 2023, 21(8): 1528-1541. |
| 32 | 杜兵帅, 邹昕蕙, 王子豪, 等. 油茶SWEET基因家族的全基因组鉴定及表达分析 [J]. 生物技术通报, 2024, 40(5): 179-190. |
| Du BS, Zou XH, Wang ZH, et al. Genome-wide identification and expression analysis of the SWEET gene family in Camellia oleifera [J]. Biotechnol Bull, 2024, 40(5): 179-190. | |
| 33 | 赵奇, 茹京娜, 李宜统, 等. 小麦Lhc基因家族鉴定与表达模式分析 [J]. 植物遗传资源学报, 2022, 23(6): 1766-1781. |
| Zhao Q, Ru JN, Li YT, et al. Identification and expression pattern analysis of Lhc gene family members in wheat [J]. J Plant Genet Resour, 2022, 23(6): 1766-1781. | |
| 34 | Qu JJ, Liu LL, Guo ZX, et al. The ubiquitous position effect, synergistic effect of recent generated tandem duplicated genes in grapevine, and their co-response and overactivity to biotic stress [J]. Fruit Res, 2023, 3(1). |
| 35 | Yu LY, Fei C, Wang DS, et al. Genome-wide identification, evolution and expression profiles analysis of bHLH gene family in Castanea mollissima [J]. Front Genet, 2023, 14: 1193953. |
| [1] | 颜伟, 陈慧婷, 叶青, 刘广超, 刘新, 侯丽霞. 葡萄HCT基因家族鉴定及其对低温胁迫的响应[J]. 生物技术通报, 2025, 41(2): 175-186. |
| [2] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [3] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| [4] | 杜品廷, 吴国江, 王振国, 李岩, 周伟, 周亚星. 高粱CPP基因家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 132-142. |
| [5] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [6] | 王子傲, 田瑞, 崔永梅, 白羿雄, 姚晓华, 安立昆, 吴昆仑. 青稞HvnJAZ4的生物信息学和表达模式分析[J]. 生物技术通报, 2025, 41(1): 173-185. |
| [7] | 李彩霞, 李艺, 穆宏秀, 林俊轩, 白龙强, 孙美华, 苗妍秀. 中国南瓜bHLH转录因子家族的鉴定与生物信息学分析[J]. 生物技术通报, 2025, 41(1): 186-197. |
| [8] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [9] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [10] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [11] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [12] | 谭博文, 张懿, 张鹏, 王振宇, 马秋香. 木薯镁离子转运蛋白家族基因的鉴定及生物信息学分析[J]. 生物技术通报, 2024, 40(9): 20-32. |
| [13] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [14] | 吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91. |
| [15] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||