








生物技术通报 ›› 2025, Vol. 41 ›› Issue (8): 124-136.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1113
• 研究报告 • 上一篇
朱丽娟( ), 张锴, 温晓蕾, 褚佳豪, 史凤玉, 王艳丽(
), 张锴, 温晓蕾, 褚佳豪, 史凤玉, 王艳丽( )
)
收稿日期:2024-11-14
出版日期:2025-08-26
发布日期:2025-07-17
通讯作者:
王艳丽,女,博士,讲师,研究方向:作物分子育种 ;E-mail: yanliwang0720@163.com作者简介:朱丽娟,女,硕士研究生,研究方向:大豆分子育种 ;E-mail: lijuanzhu91@163.com
基金资助:
ZHU Li-juan( ), ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li(
), ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li( )
)
Received:2024-11-14
Published:2025-08-26
Online:2025-07-17
摘要:
目的 野生大豆具有耐逆境特性,逐渐成为改良栽培大豆的种质资源,明确野生大豆耐镉分子调控机制,为培育耐性大豆品种提供依据。 方法 以冀东地区200份野生大豆为试验材料,利用含有75 mol/L CdCl2的Hoagland营养液处理野生大豆幼苗,并测定幼苗干重,分别对镉处理24和48 h的耐镉野生大豆R、敏感材料S进行转录组测序,对差异表达基因进行KEGG和GO富集分析,利用加权基因共表达网络分析挖掘耐镉核心基因。 结果 200份野生大豆幼苗受到不同程度镉的胁迫,与对照相比,镉处理条件下,幼苗地上部干重、地下部干重明显下降。转录组分析结果显示,在R和S材料中分别鉴定到6 443和4 496个差异表达基因。经GO和KEGG分析,发现这些差异表达基因富集到光合作用、逆境响应途径。结合加权基因共表达网络分析,挖掘到参与调控野生大豆耐镉的turquoise和blue关键模块与野生大豆耐镉性状显著相关。根据模块内基因的连接度和基因功能注释,预测LOC114376469、LOC114412091、LOC114388638、LOC114399512等8个基因可能在野生大豆镉胁迫过程中发挥作用。 结论 鉴定了2个与野生大豆耐镉相关的特异性模块,筛选到LOC114376469、LOC114412091、LOC114388638、LOC114399512等与野生大豆耐镉相关的核心基因。
朱丽娟, 张锴, 温晓蕾, 褚佳豪, 史凤玉, 王艳丽. 基于WGCNA挖掘野生大豆耐镉关键基因[J]. 生物技术通报, 2025, 41(8): 124-136.
ZHU Li-juan, ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li. Mining the Core Genes Being Tolerant to Cadmium in Wild Soybean by WGCNA[J]. Biotechnology Bulletin, 2025, 41(8): 124-136.
性状 Trait | 全距 Range | 最小值 Min | 最大值 Max | 平均值 Average | 标准差 Standard deviation |
|---|---|---|---|---|---|
| SDW-CK1 (g) | 0.090 | 0.006 | 0.096 | 0.017 | 0.008 |
| SDW-Cd1 (g) | 0.049 | 0.001 | 0.050 | 0.013 | 0.005 |
| RDW-CK1 (g) | 0.043 | 0.002 | 0.045 | 0.007 | 0.005 |
| RDW-Cd1 (g) | 0.029 | 0.001 | 0.030 | 0.005 | 0.003 |
| RSR-CK1 | 1.265 | 0.149 | 1.414 | 0.373 | 0.151 |
| RSR-Cd1 | 2.037 | 0.077 | 2.114 | 0.404 | 0.225 |
| CTC-SDW1 | 1.643 | 0.199 | 1.842 | 0.802 | 0.212 |
| CTC-RDW1 | 2.363 | 0.146 | 2.509 | 0.848 | 0.315 |
| CTC-RSR1 | 3.332 | 0.165 | 3.497 | 1.122 | 0.467 |
| SDW-CK2 (g) | 0.082 | 0.007 | 0.089 | 0.016 | 0.008 |
| SDW-Cd2 (g) | 0.059 | 0.002 | 0.061 | 0.014 | 0.007 |
| RDW-CK2 (g) | 0.056 | 0.002 | 0.058 | 0.006 | 0.005 |
| RDW-Cd2 (g) | 0.031 | 0.001 | 0.032 | 0.005 | 0.003 |
| RSR-CK2 | 1.411 | 0.158 | 1.569 | 0.364 | 0.167 |
| RSR-Cd2 | 1.180 | 0.062 | 1.242 | 0.365 | 0.165 |
| CTC-SDW2 | 3.570 | 0.166 | 3.736 | 0.877 | 0.438 |
| CTC-RDW2 | 2.500 | 0.014 | 2.514 | 0.856 | 0.342 |
| CTC-RSR2 | 2.824 | 0.039 | 2.863 | 1.068 | 0.428 |
| D1 | 1.05 | 0.14 | 1.19 | 0.459 | 0.147 |
| D2 | 1.14 | 0.02 | 1.16 | 0.512 | 0.181 |
表1 镉胁迫下野生大豆幼苗性状描述性分析
Table 1 Description analysis of wild soybean seedlings under Cd treatment
性状 Trait | 全距 Range | 最小值 Min | 最大值 Max | 平均值 Average | 标准差 Standard deviation |
|---|---|---|---|---|---|
| SDW-CK1 (g) | 0.090 | 0.006 | 0.096 | 0.017 | 0.008 |
| SDW-Cd1 (g) | 0.049 | 0.001 | 0.050 | 0.013 | 0.005 |
| RDW-CK1 (g) | 0.043 | 0.002 | 0.045 | 0.007 | 0.005 |
| RDW-Cd1 (g) | 0.029 | 0.001 | 0.030 | 0.005 | 0.003 |
| RSR-CK1 | 1.265 | 0.149 | 1.414 | 0.373 | 0.151 |
| RSR-Cd1 | 2.037 | 0.077 | 2.114 | 0.404 | 0.225 |
| CTC-SDW1 | 1.643 | 0.199 | 1.842 | 0.802 | 0.212 |
| CTC-RDW1 | 2.363 | 0.146 | 2.509 | 0.848 | 0.315 |
| CTC-RSR1 | 3.332 | 0.165 | 3.497 | 1.122 | 0.467 |
| SDW-CK2 (g) | 0.082 | 0.007 | 0.089 | 0.016 | 0.008 |
| SDW-Cd2 (g) | 0.059 | 0.002 | 0.061 | 0.014 | 0.007 |
| RDW-CK2 (g) | 0.056 | 0.002 | 0.058 | 0.006 | 0.005 |
| RDW-Cd2 (g) | 0.031 | 0.001 | 0.032 | 0.005 | 0.003 |
| RSR-CK2 | 1.411 | 0.158 | 1.569 | 0.364 | 0.167 |
| RSR-Cd2 | 1.180 | 0.062 | 1.242 | 0.365 | 0.165 |
| CTC-SDW2 | 3.570 | 0.166 | 3.736 | 0.877 | 0.438 |
| CTC-RDW2 | 2.500 | 0.014 | 2.514 | 0.856 | 0.342 |
| CTC-RSR2 | 2.824 | 0.039 | 2.863 | 1.068 | 0.428 |
| D1 | 1.05 | 0.14 | 1.19 | 0.459 | 0.147 |
| D2 | 1.14 | 0.02 | 1.16 | 0.512 | 0.181 |
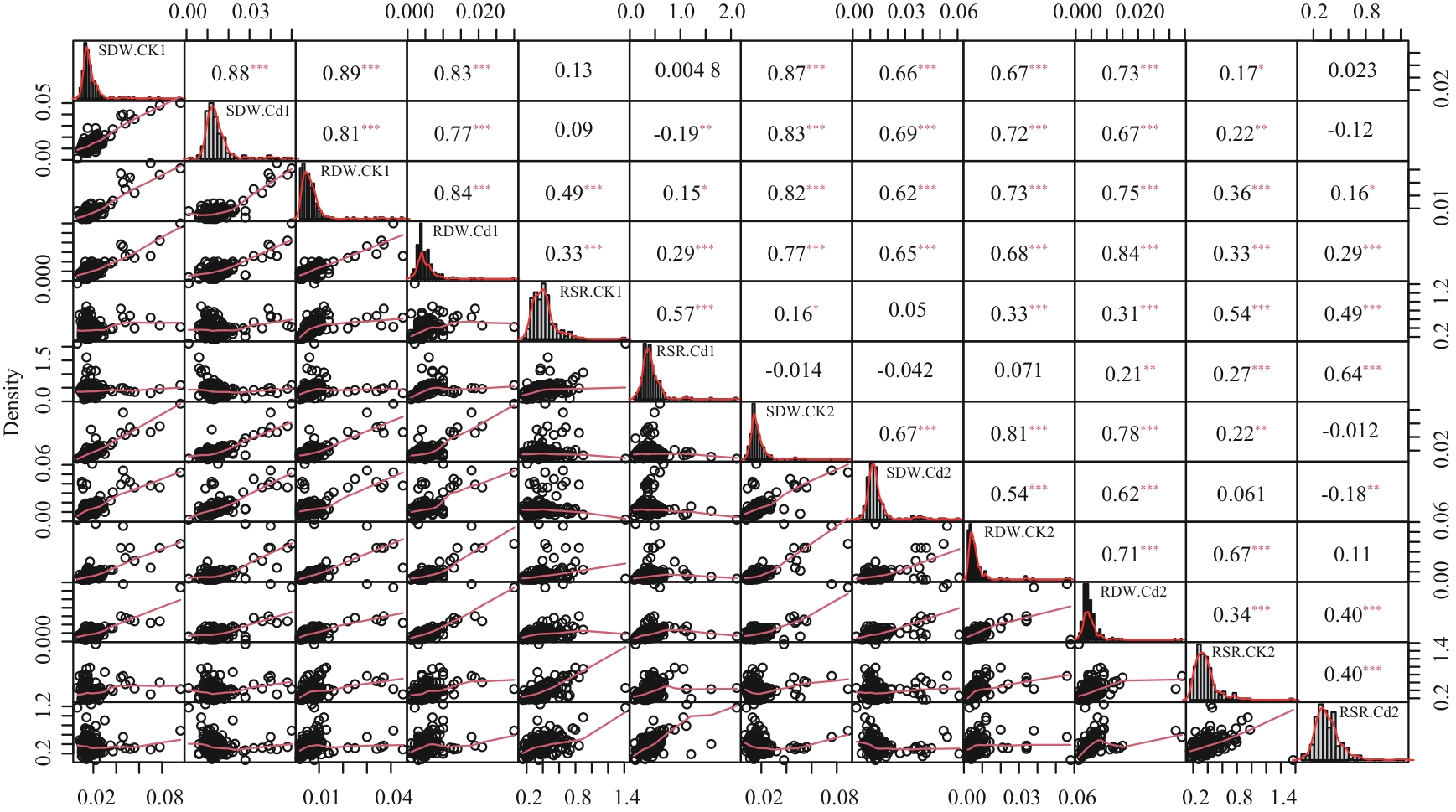
图1 两次重复下野生大豆幼苗干重相关性分析*、**和***分别表示在0.05、0.01和0.001水平上差异显著
Fig. 1 Correlation analysis of dry weight of wild soybean seedlings under two repetitions*, ** and *** indicate extremely significant correlaton at 0.05, 0.01 and 0.001 levels, respectively

图3 转录组测序数据分析A:主成分分析;B:相关性检验;C:差异表达基因热图。R0、R24、R48、S0、S24、S48代表R和S材料镉胁迫0、24和48 h。下同
Fig. 3 Analysis of transcriptome sequencing dataA: PCA analysis. B: Correlation test among samples. C: Heat map of DEGs. R0, R24, R48, S0, S24, and S48 indicate 0, 24, and 48 h under Cd stress in R and S. The same below

图4 镉胁迫下野生大豆R和S差异表达基因分析A:R和S材料不同时间点差异表达基因数量;B:R在不同时间点差异表达基因韦恩图;C:S在不同时间点差异表达基因韦恩图;D:R和S材料胁迫24 h差异表达基因韦恩图;E:R材料和S材料胁迫48 h差异表达基因韦恩图
Fig. 4 Analysis of DEGs of R and S in wild soybean under cadmium stressA: Number of DEGs. B: Venn diagram of DEGs in R at different time. C: Venn diagram of DEGs in S at different time. D: Venn diagram of DEGs in R and S at 24 h under treatment. E: Venn diagram of DEGs in R and S at 48 h under treatment

图5 GO富集分析A、B:R材料在胁迫24 h(A)、48 h(B)的差异表达基因GO富集分析;C、D:S材料在胁迫24 h(C)、48 h(D)的差异表达基因GO富集分析
Fig. 5 GO enrichment analysisA, B: GO enrichment analysis of DEGs in R at 24 h (A) and 48 h (B). C, D: GO enrichment analysis of DEGs in S at 24 h (C) and 48 h (D)
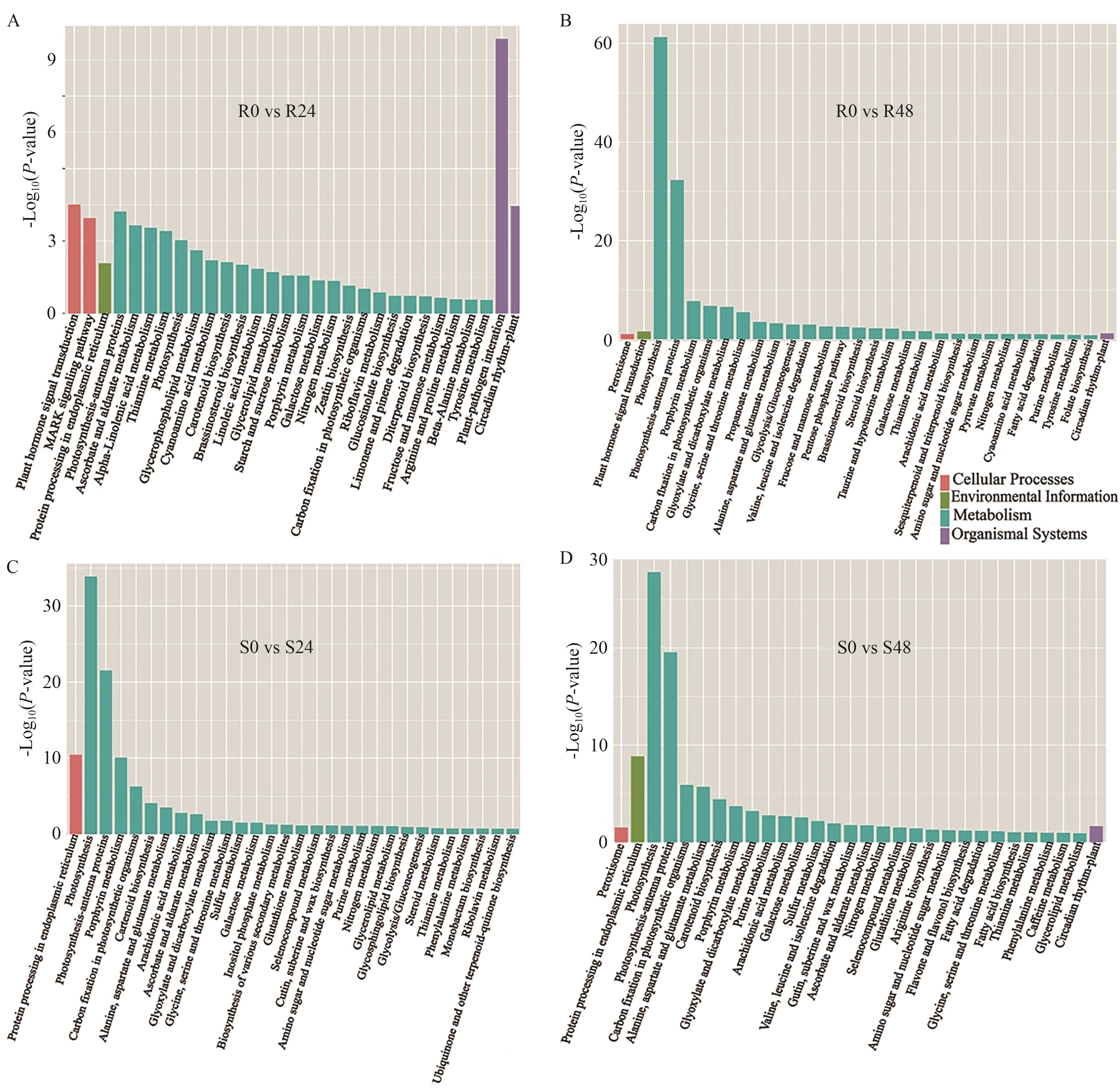
图6 KEGG富集分析A、B:R材料在胁迫24 h(A)和48 h(B)差异表达基因KEGG富集分析;C、D:S材料在胁迫24 h(C)和48 h(D)差异表达基因KEGG富集分析
Fig. 6 KEGG enrichment analysiA, B: KEGG enrichment analysis of DEGs in R at 24 h (A) and 48 h (B) under stress. C, D: KEGG enrichment analysis of DEGs in S at 24 h (C) and 48 h (D) under stress

图7 镉胁迫下野生大豆差异表达基因的WGCNA分析A:共表达模块划分;B:表型与基因模块相关性;C:blue模块共表达调控网络;D:turquoise模块共表达调控网络
Fig. 7 WGCNA enrichment analysis of DEGs in wild soybean under Cd stressA: Co-expression module division. B: Gene module eigenvalue connectivity heat map. C: Blue module co-expresses the regulatory network. D: Turquoise module co-expresses the regulatory network
关键基因 Gene ID | 基因功能注释 Gene function | 同源基因 Orthologs |
|---|---|---|
| LOC114375809 | Glutamine synthetase leaf isozyme, chloroplastic-like | Glyma 13G210800v4 |
| LOC114376496 | Protein DETOXIFICATION 42-like | Glyma 13G339800v4 |
| LOC114377985 | Stromal 70 kD heat shock-related protein, chloroplastic-like | Glyma 12G166200v4 |
| LOC114387053 | Phosphoglycerate kinase, cytosolic-like | Glyma 15G261900v4 |
| LOC114388638 | Heat shock 70 kD protein 14-like | Glyma 15G014400v4 |
| LOC114399512 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 19G139300v4 |
| LOC114406715 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 03G137000v4 |
| LOC114412091 | FAM10 family protein At4g22670-like | Glyma 05G051600v4 |
表2 八个WGCNA关键基因的功能注释
Table 2 Functional annotation of 8 WGCNA hub genes
关键基因 Gene ID | 基因功能注释 Gene function | 同源基因 Orthologs |
|---|---|---|
| LOC114375809 | Glutamine synthetase leaf isozyme, chloroplastic-like | Glyma 13G210800v4 |
| LOC114376496 | Protein DETOXIFICATION 42-like | Glyma 13G339800v4 |
| LOC114377985 | Stromal 70 kD heat shock-related protein, chloroplastic-like | Glyma 12G166200v4 |
| LOC114387053 | Phosphoglycerate kinase, cytosolic-like | Glyma 15G261900v4 |
| LOC114388638 | Heat shock 70 kD protein 14-like | Glyma 15G014400v4 |
| LOC114399512 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 19G139300v4 |
| LOC114406715 | Magnesium-chelatase subunit ChlH, chloroplastic-like | Glyma 03G137000v4 |
| LOC114412091 | FAM10 family protein At4g22670-like | Glyma 05G051600v4 |
| [1] | Mubeen S, Ni WJ, He CT, et al. Agricultural strategies to reduce cadmium accumulation in crops for food safety [J]. Agriculture, 2023, 13(2): 471. |
| [2] | Yan HL, Hezifan Z, Hao SN, et al. Cadmium contamination in food crops: risk assessment and control in smart age [J]. Crit Rev Environ Sci Technol, 2023, 53(18): 1643-1661. |
| [3] | Genchi G, Sinicropi MS, Lauria G, et al. The effects of cadmium toxicity [J]. Int J Environ Res Public Health, 2020, 17(11): 3782. |
| [4] | Finger-Teixeira A, de Lourdes Lucio Ferrarese M, Ricardo Soares A, et al. Cadmium-induced lignification restricts soybean root growth [J]. Ecotoxicol Environ Saf, 2010, 73(8): 1959-1964. |
| [5] | He SL, Wang YS, Li DZ, et al. Environmental and historical determinants of patterns of genetic differentiation in wild soybean (Glycine soja sieb. et zucc) [J]. Sci Rep, 2016, 6: 22795. |
| [6] | Wang KJ, Li F, Ali Cheema A. Studies on the distribution of wild soybean (Glycine soja) in China [J]. Pak J Biol Sci, 2001, 4(2): 149-155. |
| [7] | 李明霞. 盐和低氮胁迫下栽培大豆和野大豆适应性比较研究 [D]. 长春: 东北师范大学, 2020. |
| Li MX. Comparative study on adaptability of cultivated soybean and wild soybean under salt and low nitrogen stress [D]. Changchun: Northeast Normal University, 2020. | |
| [8] | Liu DP, Li MX, Liu Y, et al. Integration of the metabolome and transcriptome reveals the resistance mechanism to low nitrogen in wild soybean seedling roots [J]. Environ Exp Bot, 2020, 175: 104043. |
| [9] | 赵忠娟, 魏艳丽, 李哲, 等. 野大豆与栽培大豆愈伤组织耐盐性比较 [J]. 山东科学, 2015, 28(1): 102-108. |
| Zhao ZJ, Wei YL, Li Z, et al. Salinity tolerance comparison for the calli of wild and cultivated soybeans [J]. Shandong Sci, 2015, 28(1): 102-108. | |
| [10] | 符辉. 干旱胁迫下野大豆(Glycine soja sieb.et zucc.)和大豆(Glycine max L.)幼苗叶片代谢组学比较研究 [D]. 长春: 东北师范大学, 2020. |
| Fu H. Comparative study on leaf metabonomics of Glycine soja sieb.et zucc. and Glycine max L. seedlings under drought stress [D]. Changchun: Northeast Normal University, 2020. | |
| [11] | Gao Y, Tao B, Qiu LJ, et al. Role of physiological mechanisms and EPSPS gene expression in glyphosate resistance in wild soybeans (Glycine soja) [J]. Pestic Biochem Physiol, 2014, 109: 6-11. |
| [12] | 袁翠平, 齐广勋, 李玉秋, 等. 野生大豆抗胞囊线虫QTL定位 [J]. 中国油料作物学报, 2019, 41(6): 887-893. |
| Yuan CP, Qi GX, Li YQ, et al. QTL mapping for resistance to soybean cyst nematode in wild soybean [J]. Chin J Oil Crop Sci, 2019, 41(6): 887-893. | |
| [13] | 陈爱国, 王岩, 孟未来, 等. 不同原生境来源野生大豆抗花叶病毒(SMV)综合评价及聚类分析 [J]. 辽宁农业科学, 2020(1): 7-13. |
| Chen AG, Wang Y, Meng WL, et al. Evaluation, cluster analysis for Glycine soja of different habitats resistant to soybean mosaic virus (SMV) [J]. Liaoning Agric Sci, 2020(1): 7-13. | |
| [14] | 陈珊宇, 王大刚, 郑桂杰, 等. 野生大豆对大豆花叶病毒株系SC13的抗性遗传和基因定位 [J]. 植物遗传资源学报, 2020, 21(1): 139-145. |
| Chen SY, Wang DG, Zheng GJ, et al. Inheritance and gene mapping of resistance to soybean mosaic virus strain SC13 in soybean [Glycine soja sieb. & zucc.] [J]. J Plant Genet Resour, 2020, 21(1): 139-145. | |
| [15] | Arao T, Ae N, Sugiyama MT, et al. Genotypic differences in cadmium uptake and distribution in soybeans [J]. Plant Soil, 2003, 251(2): 247-253. |
| [16] | 赵毅, 于翠梅, 杨柳, 等. 野生大豆和不同栽培大豆品种在镉胁迫下种子萌发及幼苗生长的差异 [J]. 大豆科学, 2019, 38(2): 267-273. |
| Zhao Y, Yu CM, Yang L, et al. Differences of cadmium stress on seed germination and seedling growth in the wild soybean and cultivated soybeans [J]. Soybean Sci, 2019, 38(2): 267-273. | |
| [17] | 冯君, 赵毅, 高婷, 等. 野生和栽培大豆对镉胁迫的响应差异分析 [J]. 大豆科学, 2018, 37(5): 756-761. |
| Feng J, Zhao Y, Gao T, et al. The difference of responses to the cadmium stress between a wild soybean and a cultivated soybean [J]. Soybean Sci, 2018, 37(5): 756-761. | |
| [18] | Yu HW, Yang ZM, Wang JF, et al. Identification of key genes and metabolites involved in meat quality performance in Qinchuan cattle by WGCNA [J]. J Integr Agric, 2024, 23(11): 3923-3937. |
| [19] | 夏雪岩, 崔纪菡, 黄玫红, 等. 谷子苗期氮高效转录组分析与基因挖掘 [J]. 中国农业科技导报, 2024, 26(10): 41-57. |
| Xia XY, Cui JH, Huang MH, et al. Analysis of high-efficiency transcriptome of nitrogen in millet seedlings and gene mining [J]. J Agric Sci Technol, 2024, 26(10): 41-57. | |
| [20] | 陈晓涓, 王海菊, 王富敏, 等. 基于WGCNA鉴定全缘叶绿绒蒿类黄酮合成途径关键基因 [J]. 中国农业科学, 2024, 57(15): 3053-3070. |
| Chen XJ, Wang HJ, Wang FM, et al. Identification of key genes in the flavonoid synthesis pathway of Meconopsis integrifolia based on WGCNA [J]. Sci Agric Sin, 2024, 57(15): 3053-3070. | |
| [21] | 张会, 王越越, 赵波, 等. 基于WGCNA的谷子苗期冷胁迫应答基因网络构建与核心因子发掘 [J]. 中国农业科技导报, 2023, 25(10): 22-34. |
| Zhang H, Wang YY, Zhao B, et al. Identification of co-expression genes related to cold stress in foxtail millet by WGCNA [J]. J Agric Sci Technol, 2023, 25(10): 22-34. | |
| [22] | 周雨青, 杨永飞, 葛常伟, 等. 基于WGCNA的棉花子叶抗冷相关共表达模块鉴定 [J]. 中国农业科技导报, 2022, 24(4): 52-62. |
| Zhou YQ, Yang YF, Ge CW, et al. Identification of cold-related co-expression modules in cotton Cotyledon by WGCNA [J]. J Agric Sci Technol, 2022, 24(4): 52-62. | |
| [23] | 王世瑶. 低氮胁迫下不同生态型野大豆(Glycine soja)新老叶片光合特性,离子平衡和氮代谢的研究 [D]. 长春: 东北师范大学, 2021. |
| Wang SY. Adaptive variations for nitrogen deficiency in photosynthetic characteristics, ion balance and nitrogen metabolism in young and old leaves of different ecotypes wild soybean[D]. Changchun: Northeast Normal University, 2021. | |
| [24] | Liu XL, Zhang CL, Lamlom SF, et al. Genetic adaptations of soybean to cold stress reveal key insights through transcriptomic analysis [J]. Biology, 2024, 13(11): 856. |
| [25] | Cheng Y, Cheng XQ, Wei K, et al. Comparative transcriptome analysis of salt-tolerant and-sensitive soybean cultivars under salt stress [J]. Int J Mol Sci, 2024, 25(18): 9818. |
| [26] | Li MQ, Li HN, Sun AN, et al. Transcriptome analysis reveals key drought-stress-responsive genes in soybean [J]. Front Genet, 2022, 13: 1060529. |
| [27] | Zheng SL, Qi J, Fu TW, et al. Novel mechanisms of cadmium tolerance and Cd-induced fungal stress in wheat: Transcriptomic and metagenomic insights [J]. Ecotoxicol Environ Saf, 2023, 256: 114842. |
| [28] | 罗玲, 许肖恒, 杨康, 等. 非生物胁迫下植物衰老和热激蛋白响应 [J]. 草业科学, 2020, 37(11): 2320-2333. |
| Luo L, Xu XH, Yang K, et al. Senescence and heat shock protein in plants in response to abiotic stress [J]. Pratacultural Sci, 2020, 37(11): 2320-2333. | |
| [29] | Luo JS, Zhang ZH. Mechanisms of cadmium phytoremediation and detoxification in plants [J]. Crop J, 2021, 9(3): 521-529. |
| [30] | Feng Z, Ji SY, Ping JF, et al. Recent advances in metabolomics for studying heavy metal stress in plants [J]. Trac Trends Anal Chem, 2021, 143: 116402. |
| [31] | Ma SC, Lapin D, Liu L, et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme [J]. Science, 2020, 370(6521): eabe3069. |
| [1] | 郭秀娟, 冯宇, 吴瑞香, 王利琴, 杨建春. Ca2+处理对胡麻种子萌发影响的转录组分析[J]. 生物技术通报, 2025, 41(7): 139-149. |
| [2] | 冯冰, 闫彩霞, 刘艺, 董凯悦, 赵楠, 赵瑞, 陈少良. 灰杨PcAHL17负调控拟南芥的镉耐受性[J]. 生物技术通报, 2025, 41(6): 269-283. |
| [3] | 周志国, 樊双虎, 邓晨, 冯雪. 2,4-表油菜素内酯对镉胁迫下胡萝卜幼苗生理特性的影响[J]. 生物技术通报, 2025, 41(5): 165-174. |
| [4] | 王斌, 王玉昆, 肖艳辉. 丁香罗勒(Ocimum gratissimum)叶片响应镉胁迫的比较转录组学分析[J]. 生物技术通报, 2025, 41(3): 255-270. |
| [5] | 岳丽昕, 王清华, 刘泽洲, 孔素萍, 高莉敏. 基于转录组和WGCNA筛选大葱雄性不育相关基因[J]. 生物技术通报, 2024, 40(9): 212-224. |
| [6] | 文洁, 杜元欣, 吴安波, 杨广容, 鲁敏, 安华明, 南红. 刺梨SOD基因家族鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(5): 153-166. |
| [7] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [8] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [9] | 姜南, 石杨, 赵志慧, 李斌, 赵熠辉, 杨俊彪, 闫家铭, 靳雨璠, 陈稷, 黄进. 镉胁迫下水稻OsPT1的表达及功能分析[J]. 生物技术通报, 2023, 39(1): 166-174. |
| [10] | 呼艳姣, 陈美凤, 强瑀, 李海燕, 刘静, 秦樊鑫. 镉胁迫下锌硒交互作用对水稻镉毒害的缓解机制[J]. 生物技术通报, 2022, 38(4): 143-152. |
| [11] | 祖国蔷, 胡哲, 王琪, 李光哲, 郝林. Burkholderia sp. GD17对水稻幼苗镉耐受的调节[J]. 生物技术通报, 2022, 38(4): 153-162. |
| [12] | 杨馥榕, 王晓红, 肖琪, 方娟, 李立华. 木槿品种对镉胁迫的生理响应及耐镉能力评价[J]. 生物技术通报, 2022, 38(1): 98-107. |
| [13] | 王竹承, 刘辉, 李荣华. 外源硫对镉胁迫下马齿苋光合性状和矿质元素吸收的影响[J]. 生物技术通报, 2020, 36(3): 133-140. |
| [14] | 王竹承, 刘辉, 李荣华, 陈新, 李欣, 路致远. 外源硫与乙烯缓解马齿苋镉胁迫的生理机制研究[J]. 生物技术通报, 2019, 35(10): 71-79. |
| [15] | 马晓丽, 冀瑞萍, 田保华, 霍建新. 一氧化氮(NO)对镉胁迫下小麦幼苗氧化损伤的影响[J]. 生物技术通报, 2017, 33(5): 102-107. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||