








生物技术通报 ›› 2022, Vol. 38 ›› Issue (12): 204-213.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0330
孙燕1,2( ), 王金刚1(
), 王金刚1( ), 臧丹丹2(
), 臧丹丹2( ), 赵恒田2, 刘淑华2
), 赵恒田2, 刘淑华2
收稿日期:2022-03-19
出版日期:2022-12-26
发布日期:2022-12-29
作者简介:孙燕,女,硕士研究生,研究方向:园林植物与应用;E-mail:基金资助:
SUN Yan1,2( ), WANG Jin-gang1(
), WANG Jin-gang1( ), ZANG Dan-dan2(
), ZANG Dan-dan2( ), ZHAO Heng-tian2, LIU Shu-hua2
), ZHAO Heng-tian2, LIU Shu-hua2
Received:2022-03-19
Published:2022-12-26
Online:2022-12-29
摘要:
为研究不同发育时期(绿熟期和成熟期)蓝果忍冬果实基因的表达差异,探索蓝果忍冬在生长发育过程中的关键代谢通路,以‘中科蓝1号’蓝果忍冬品种为研究对象,利用高通量测序技术对蓝果忍冬绿熟期和成熟期果实的转录组数据进行比较分析。结果表明,共有40.32 Gb Clean Data,每个样品Clean Data均大于6.07 Gb,Q30碱基百分比均在93.89%及以上,GC含量为46.32%-49.38%;两个发育时期的果实共检测出3 247 个差异表达基因,包括1 642 个上调基因和1 605 个下调基因;GO功能富集分析发现,蓝果忍冬果实发育过程中的关键调控基因主要参与代谢、细胞和细胞组分以及催化过程;KEGG富集分析发现,差异表达基因主要富集在植物激素信号转导、碳代谢和氨基酸生物合成等途径。该结果为进一步研究蓝果忍冬果实发育过程中的动态变化提供了参考,为后续开展蓝果忍冬功能基因鉴定、遗传多样性分析等研究提供理论依据。
孙燕, 王金刚, 臧丹丹, 赵恒田, 刘淑华. 不同发育时期蓝果忍冬果实的转录组分析[J]. 生物技术通报, 2022, 38(12): 204-213.
SUN Yan, WANG Jin-gang, ZANG Dan-dan, ZHAO Heng-tian, LIU Shu-hua. Transcriptome Analysis of Lonicera caerulea Fruits at Different Developmental Stages[J]. Biotechnology Bulletin, 2022, 38(12): 204-213.
| 基因Gene | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| OM964861 | CCCTGGGGTC | CTCAGAGGAATTGAATTC |
| OM964862 | GCAAGAACTATTTAACAG | CTCACCACCAGTTTG |
| OM964867 | GTCGAATGTGTCAAC | CATTAGATCCTGGCTGC |
| OM964864 | CGAGCTAATGGACACGGTG | GTAGCCTTTCTCTAATCC |
| OM964868 | CTATTCCGAGAGATTTG | GTTCTTTGGTTCCCATC |
| OM964869 | GGTTTACATTTTAGCGAC | CAACTTGTCCTCACCAAC |
| OM964870 | CTTTGAATCCCATTGATTC | GAATTGCTACTGGCAAGC |
| OM964871 | GCTGGTCATTTTACAG | CACTGCCCTTATTAGTG |
| OM964872 | CCACCGACCTTCCC | GACATGTTCCGCGCTC |
| OM964873 | CAACGATTTCCGAAC | CCGGTGTCACCTTTTCC |
| MT344113 | CGTCGTTTGTGACAATGG | GGTCCCACACATGACCC |
| MT344114 | GTCTGTGCCGAACACGGC | CAGTATAATGGCCTTTAGC |
表1 转录组数据RT-qPCR验证引物序列
Table 1 Primer sequences used in RT-qPCR validation of transcriptome data
| 基因Gene | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| OM964861 | CCCTGGGGTC | CTCAGAGGAATTGAATTC |
| OM964862 | GCAAGAACTATTTAACAG | CTCACCACCAGTTTG |
| OM964867 | GTCGAATGTGTCAAC | CATTAGATCCTGGCTGC |
| OM964864 | CGAGCTAATGGACACGGTG | GTAGCCTTTCTCTAATCC |
| OM964868 | CTATTCCGAGAGATTTG | GTTCTTTGGTTCCCATC |
| OM964869 | GGTTTACATTTTAGCGAC | CAACTTGTCCTCACCAAC |
| OM964870 | CTTTGAATCCCATTGATTC | GAATTGCTACTGGCAAGC |
| OM964871 | GCTGGTCATTTTACAG | CACTGCCCTTATTAGTG |
| OM964872 | CCACCGACCTTCCC | GACATGTTCCGCGCTC |
| OM964873 | CAACGATTTCCGAAC | CCGGTGTCACCTTTTCC |
| MT344113 | CGTCGTTTGTGACAATGG | GGTCCCACACATGACCC |
| MT344114 | GTCTGTGCCGAACACGGC | CAGTATAATGGCCTTTAGC |
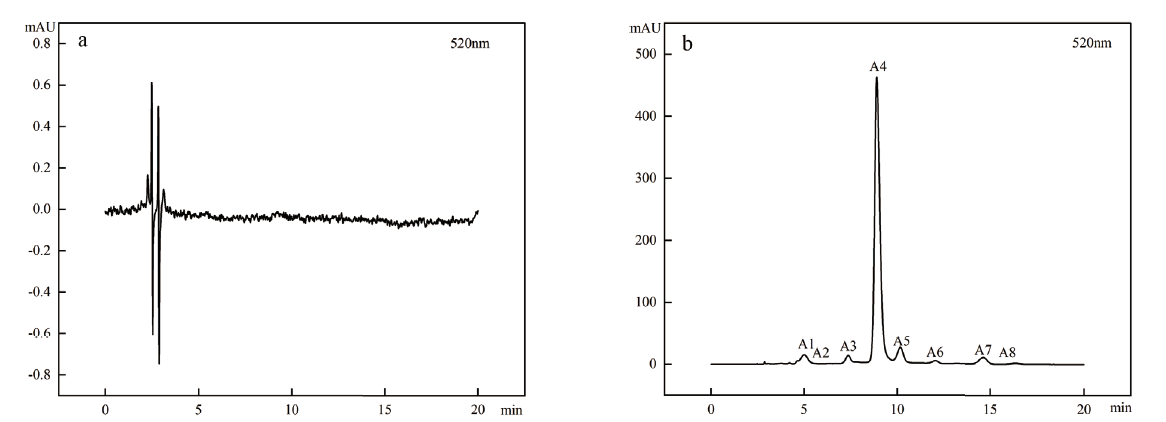
图2 ‘中科蓝1号’不同发育时期果实高效液相色谱 a:蓝果忍冬绿熟期;b:蓝果忍冬成熟期;A1:矢车菊素-3,5-双葡萄糖苷;A2:飞燕草素-3-O-葡萄糖苷;A3:飞燕草素-3-O-芸香糖苷;A4:矢车菊素-3-O-葡萄糖苷;A5:矢车菊素-3-O-芸香糖苷;A6:天竺葵素-3-O-葡萄糖苷;A7:芍药素-3-O-葡萄糖苷;A8:芍药素-3-O-芸香糖苷
Fig. 2 HPLC of ‘Zhongkelan No.1’ fruit at different developmental stages a:Green ripening stage of Lonicera caerulea fruit. b:Ripening stage of Lonicera caerulea fruit. A1:Cyanidin-3,5-diglucoside. A2:Delphinidin-3-O-glucoside. A3:Delphinidin-3-O-rutinosidchlorid. A4:Cyanidin-3-O-glucoside. A5:Cyanidin-3-O-rutinoside. A6:Pelargonidin-3-O-glucoside. A7:Peonidin-3-O-glucoside. A8:Peonidin-3-O-rutinoside
| Sample | Clean reads | Clean bases | GC Content/% | Q30/% |
|---|---|---|---|---|
| B01 | 23 970 275 | 7 147 808 408 | 46.32 | 94.50 |
| B02 | 21 515 272 | 6 404 727 658 | 47.02 | 94.18 |
| B03 | 21 364 950 | 6 366 085 640 | 46.64 | 93.89 |
| G01 | 20 399 412 | 6 074 481 494 | 49.24 | 94.10 |
| G02 | 22 320 706 | 6 652 355 802 | 48.67 | 94.26 |
| G03 | 25 781 714 | 7 672 240 582 | 49.38 | 93.90 |
表3 测序数据统计结果
Table 3 Statistics of sequencing data
| Sample | Clean reads | Clean bases | GC Content/% | Q30/% |
|---|---|---|---|---|
| B01 | 23 970 275 | 7 147 808 408 | 46.32 | 94.50 |
| B02 | 21 515 272 | 6 404 727 658 | 47.02 | 94.18 |
| B03 | 21 364 950 | 6 366 085 640 | 46.64 | 93.89 |
| G01 | 20 399 412 | 6 074 481 494 | 49.24 | 94.10 |
| G02 | 22 320 706 | 6 652 355 802 | 48.67 | 94.26 |
| G03 | 25 781 714 | 7 672 240 582 | 49.38 | 93.90 |
| 不同长度区间 Length range/bp | 转录本序列数量 Transcripts | 单基因序列数量 Unigene |
|---|---|---|
| 200-300 | 37 818(16.02%) | 29 441(36.00%) |
| 300-500 | 34 253(14.51%) | 20 582(25.17%) |
| 500-1 000 | 44 975(19.05%) | 14 415(17.63%) |
| 1 000-2 000 | 60 698(25.72%) | 9 656(11.81%) |
| 2 000+ | 58 289(24.70%) | 7 683(9.40%) |
| Total number | 236 033 | 81 777 |
| Total Length | 323 833 751 | 62 424 481 |
| N50 Length | 2 186 | 1 421 |
| Mean Length | 1 371.99 | 763.35 |
表4 组装结果统计
Table 4 Assembly result statistics
| 不同长度区间 Length range/bp | 转录本序列数量 Transcripts | 单基因序列数量 Unigene |
|---|---|---|
| 200-300 | 37 818(16.02%) | 29 441(36.00%) |
| 300-500 | 34 253(14.51%) | 20 582(25.17%) |
| 500-1 000 | 44 975(19.05%) | 14 415(17.63%) |
| 1 000-2 000 | 60 698(25.72%) | 9 656(11.81%) |
| 2 000+ | 58 289(24.70%) | 7 683(9.40%) |
| Total number | 236 033 | 81 777 |
| Total Length | 323 833 751 | 62 424 481 |
| N50 Length | 2 186 | 1 421 |
| Mean Length | 1 371.99 | 763.35 |
| 数据库 Database | 注释的单基因数Number of annotated unigenes | 300 bp≤长度<1 000 bp 300 bp≤Length<1 000 bp | 长度≥1 000 bp Length≥1 000 bp |
|---|---|---|---|
| COG | 8 056 | 1 670 | 5 646 |
| GO | 17 733 | 5 300 | 9 680 |
| KEGG | 9 814 | 2 805 | 5 673 |
| KOG | 15 668 | 4 567 | 8 845 |
| Pfam | 18 933 | 5 043 | 12 068 |
| Swissprot | 19 579 | 5 863 | 11 158 |
| eggNOG | 26 275 | 8 298 | 14 187 |
| nr | 28 639 | 9 428 | 14 621 |
| In all database | 28 992 | 9 544 | 14 648 |
表5 单基因功能注释统计
Table 5 Single-gene function annotation statistics
| 数据库 Database | 注释的单基因数Number of annotated unigenes | 300 bp≤长度<1 000 bp 300 bp≤Length<1 000 bp | 长度≥1 000 bp Length≥1 000 bp |
|---|---|---|---|
| COG | 8 056 | 1 670 | 5 646 |
| GO | 17 733 | 5 300 | 9 680 |
| KEGG | 9 814 | 2 805 | 5 673 |
| KOG | 15 668 | 4 567 | 8 845 |
| Pfam | 18 933 | 5 043 | 12 068 |
| Swissprot | 19 579 | 5 863 | 11 158 |
| eggNOG | 26 275 | 8 298 | 14 187 |
| nr | 28 639 | 9 428 | 14 621 |
| In all database | 28 992 | 9 544 | 14 648 |
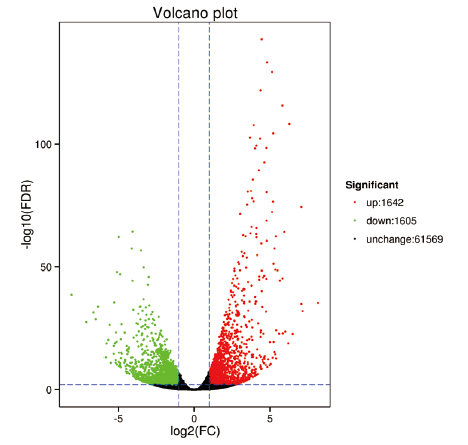
图3 差异表达基因的火山图图中绿色和红色的点代表有显著性表达差异的基因,绿色代表基因表达量下调,红色代表基因表达量上调,黑色的点代表无显著性表达差异的基因
Fig. 3 Volcano map of differentially expressed genes The green and red dots in the graph indicate genes with significant expression differences,green indicates genes with down-regulated expression,red indicates genes with up-regulated expression,and black dots indicate genes without significant expression differences
| [1] | Plekhanova MN. Blue honeysuckle(Lonicera caerulea L.)- a new commercial berry crop for temperate climate:genetic resources and breeding[J]. Acta Hortic, 2000(538):159-164. |
| [2] | 霍俊伟, 杨国慧, 睢薇, 等. 蓝靛果忍冬(Lonicera caerulea)种质资源研究进展[J]. 园艺学报, 2005, 32(1):159-164. |
| Huo JW, Yang GH, Sui W, et al. Review of study on germplasm resources of blue honeysuckle(Lonicera caerulea L.)[J]. Acta Hortic Sin, 2005, 32(1):159-164. | |
| [3] |
Chaovanalikit A, Thompson MM, Wrolstad RE. Characterization and quantification of anthocyanins and polyphenolics in blue honeysuckle(Lonicera caerulea L.)[J]. J Agric Food Chem, 2004, 52(4):848-852.
doi: 10.1021/jf030509o URL |
| [4] |
Celli GB, Ghanem A, Brooks MSL. Optimization of ultrasound-assisted extraction of anthocyanins from haskap berries(Lonicera caerulea L.)using response surface methodology[J]. Ultrason Sonochem, 2015, 27:449-455.
doi: 10.1016/j.ultsonch.2015.06.014 URL |
| [5] | 刘奕琳. 蓝靛果花色苷分离及其抗氧化与抗癌功能研究[D]. 哈尔滨: 东北林业大学, 2012. |
| Liu YL. Sepaeration and reseach on antioxidant and anticancer anthocyanin of lonecera edulis[D]. Harbin: Northeast Forestry University, 2012. | |
| [6] |
Wang LS, Stoner GD. Anthocyanins and their role in cancer prevention[J]. Cancer Lett, 2008, 269(2):281-290.
doi: 10.1016/j.canlet.2008.05.020 URL |
| [7] | 张建全, 张含生, 张壮飞, 等. 小兴安岭地区蓝靛果优良品种引种栽培试验初报[J]. 农业科技通讯, 2018(1):152-155. |
| Zhang JQ, Zhang HS, Zhang ZF, et al. A preliminary report on the introduction and cultivation trials of good varieties of blue honeysuckle in the Xiaoxinganling region[J]. Bulletin of Agricultural Science and Technology, 2018(1):152-155. | |
| [8] |
Kong JM, Chia LS, Goh NK, et al. Analysis and biological activities of anthocyanins[J]. Phytochemistry, 2003, 64(5):923-933.
doi: 10.1016/S0031-9422(03)00438-2 URL |
| [9] |
Lohachoompol V, Mulholland M, Srzednicki G, et al. Determination of anthocyanins in various cultivars of highbush and rabbiteye blueberries[J]. Food Chem, 2008, 111(1):249-254.
doi: 10.1016/j.foodchem.2008.03.067 URL |
| [10] | 王忠华, 俞超, 吴月燕. 葡萄花青素苷生物合成研究进展[J]. 生物技术通报, 2012(9):1-7. |
| Wang ZH, Yu C, Wu YY. Progress on anthocyanin biosynthesis in grape[J]. Biotechnol Bull, 2012(9):1-7. | |
| [11] |
Springob K, Nakajima JI, Yamazaki M, et al. Recent advances in the biosynthesis and accumulation of anthocyanins[J]. Nat Prod Rep, 2003, 20(3):288-303.
pmid: 12828368 |
| [12] |
Zhou LP, Wang H, Yi JJ, et al. Anti-tumor properties of anthocyanins from Lonicera caerulea ‘Beilei’ fruit on human hepatocellular carcinoma:in vitro and in vivo study[J]. Biomed Pharmacother, 2018, 104:520-529.
doi: 10.1016/j.biopha.2018.05.057 URL |
| [13] | Pažereckaitė A, Jasutienė I, Šarkinas A, et al. Antimicrobial activity and composition of different cultivars of honeysuckle berry Lonicera caerulea L[C]// The 1st International Electronic Conference on Plant Science. Basel Switzerland: MDPI, 2020:1-15. |
| [14] |
Montefiori M, McGhie TK, Costa G, et al. Pigments in the fruit of red-fleshed kiwifruit(Actinidia chinensis and Actinidia deliciosa)[J]. J Agric Food Chem, 2005, 53(24):9526-9530.
doi: 10.1021/jf051629u URL |
| [15] |
Elitzur T, Yakir E, Quansah L, et al. Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security[J]. Plant Physiol, 2016, 171(1):380-391.
doi: 10.1104/pp.15.01866 pmid: 26956665 |
| [16] | 齐晓. 紫花苜蓿在转录组水平响应低温胁迫的差异表达基因研究[D]. 北京: 中国农业科学院, 2017. |
| Qi X. The analysis of the differentially expressed genes in alfalfa under cold stress at transcriptome level[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. | |
| [17] |
Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nat Biotechnol, 2011, 29(7):644-652.
doi: 10.1038/nbt.1883 pmid: 21572440 |
| [18] | 押辉远, 陈叶, 张岩松, 等. 槟榔不同发育时期果实转录组特征分析[J]. 热带作物学报, 2020, 41(7):1279-1287. |
| Ya HY, Chen Y, Zhang YS, et al. Analysis of transcriptome characteristics of Areca at different developmental stages[J]. Chin J Trop Crops, 2020, 41(7):1279-1287. | |
| [19] |
Raudsepp P, Anton D, Roasto M, et al. The antioxidative and antimicrobial properties of the blue honeysuckle(Lonicera caerulea L.), Siberian rhubarb(Rheum rhaponticum L.)and some other plants, compared to ascorbic acid and sodium nitrite[J]. Food Control, 2013, 31(1):129-135.
doi: 10.1016/j.foodcont.2012.10.007 URL |
| [20] |
Wu SS, He X, Wu XS, et al. Inhibitory effects of blue honeysuckle(Lonicera caerulea L)on adjuvant-induced arthritis in rats:Crosstalk of anti-inflammatory and antioxidant effects[J]. J Funct Foods, 2015, 17:514-523.
doi: 10.1016/j.jff.2015.06.007 URL |
| [21] | Zhao L, Li SR, Zhao L, et al. Antioxidant activities and major bioactive components of consecutive extracts from blue honeysuckle(Lonicera caerulea L.)cultivated in China[J]. J Food Biochem, 2015, 39(6):653-662. |
| [22] | 呼延文静. 蓝果忍冬种子及其籽油主要成分测定及抗氧化活性研究[D]. 哈尔滨: 东北农业大学, 2021. |
| Huyan WJ. Determination of main components and antioxidant activity of blue honeysuckle seeds and its seed oil[D]. Harbin: Northeast Agricultural University, 2021. | |
| [23] |
Huo JW, Liu P, Wang Y, et al. De novo transcriptome sequencing of blue honeysuckle fruit(Lonicera caerulea L.)and analysis of major genes involved in anthocyanin biosynthesis[J]. Acta Physiol Plant, 2016, 38(7):1-12.
doi: 10.1007/s11738-015-2023-4 URL |
| [24] | Yang Z, Chen H, Jia J, et al. De novo assembly and discovery of metabolic pathways and genes that are involved in defense against pests in Songyun Pinus massoniana Lamb[J]. Bangladesh J Bot, 2016, 45(4):855-863. |
| [25] |
Song HW, Zhao XX, Hu WC, et al. Comparative transcriptional analysis of loquat fruit identifies major signal networks involved in fruit development and ripening process[J]. Int J Mol Sci, 2016, 17(11):1837.
doi: 10.3390/ijms17111837 URL |
| [26] |
Li CQ, Wang Y, Huang XM, et al. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading[J]. BMC Genomics, 2013, 14:552.
doi: 10.1186/1471-2164-14-552 pmid: 23941440 |
| [27] | 徐伟君, 张九东, 郑博文, 等. 基于高通量测序的大叶女贞(Ligustrum lucidum)转录组分析[J]. 分子植物育种, 2019, 17(19):6295-6299. |
| Xu WJ, Zhang JD, Zheng BW, et al. Transcriptome analysis of Ligustrum lucidum based on high-throughput sequencing[J]. Mol Plant Breed, 2019, 17(19):6295-6299. | |
| [28] |
Li XY, Sun HY, Pei JB, et al. De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants[J]. Gene, 2012, 511(1):54-61.
doi: 10.1016/j.gene.2012.09.021 pmid: 22995346 |
| [29] |
赵阳阳, 郭雨潇, 张凌云. 文冠果果实转录组测序及分析[J]. 生物技术通报, 2019, 35(6):24-31.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-1101 URL |
| Zhao YY, Guo YX, Zhang LY. Transcriptome sequencing and analysis of Xanthoceras sorbifolia bunge fruit[J]. Biotechnol Bull, 2019, 35(6):24-31. | |
| [30] | 彭佳佳. 红掌转录组测序及花青素生物合成途径差异表达基因的分析[D]. 银川: 宁夏大学, 2015. |
| Peng JJ. Transcriptome analysis and anthocyanin biosynthesis genetic expression in the spathe of Anthurium andreanum[D]. Yinchuan: Ningxia University, 2015. | |
| [31] |
Dorcey E, Urbez C, Blázquez MA, et al. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis[J]. Plant J, 2009, 58(2):318-332.
doi: 10.1111/j.1365-313X.2008.03781.x URL |
| [32] |
Groot SPC, Bruinsma J, Karssen CM. The role of endogenous gibberellin in seed and fruit development of tomato:studies with a gibberellin-deficient mutant[J]. Physiol Plant, 1987, 71(2):184-190.
doi: 10.1111/j.1399-3054.1987.tb02865.x URL |
| [33] | Stopar M. KNOCHE, Moritz, KHANAL, Bischnu P., STOPAR Matej. Russeting and microcracking of ‘Golden Delicious’ apple fruit concomitantly decline due to gibberellin A4+7 application[J]. J Am Soc Hortic Sci, 2011, 136(3):159-164. |
| [34] |
Serrani JC, Fos M, Atarés A, et al. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv micro-tom of tomato[J]. J Plant Growth Regul, 2007, 26(3):211-221.
doi: 10.1007/s00344-007-9014-7 URL |
| [35] | 赵荣华, 白世践, 陈光, 等. 赤霉素对无核白葡萄果实品质的影响[J]. 黑龙江农业科学, 2014(9):40-42. |
| Zhao RH, Bai SJ, Chen G, et al. Effect of gibberellin on fruit quality of thompsons seedless[J]. Heilongjiang Agric Sci, 2014(9):40-42. | |
| [36] |
Tiwari A, Offringa R, Heuvelink E. Auxin-induced fruit set in Capsicum annuum L. requires downstream gibberellin biosynthesis[J]. J Plant Growth Regul, 2012, 31(4):570-578.
doi: 10.1007/s00344-012-9267-7 URL |
| [37] |
Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, et al. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits[J]. J Exp Bot, 2014, 65(2):401-417.
doi: 10.1093/jxb/ert377 pmid: 24277278 |
| [38] |
Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits[J]. Trends Plant Sci, 2013, 18(9):477-483.
doi: 10.1016/j.tplants.2013.06.003 pmid: 23870661 |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [3] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [4] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [5] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [6] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [7] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [8] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [9] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [10] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [11] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [12] | 孙言秋, 谢采芸, 汤岳琴. 耐高温酿酒酵母的构建与高温耐受机制解析[J]. 生物技术通报, 2023, 39(11): 226-237. |
| [13] | 徐俊, 叶雨晴, 牛雅静, 黄河, 张蒙蒙. 菊花根状茎发育的转录组分析[J]. 生物技术通报, 2023, 39(10): 231-245. |
| [14] | 罗皓天, 王龙, 王禹茜, 王月, 李佳祯, 杨梦珂, 张杰, 邓欣, 王红艳. 青狗尾草RNAi途径相关基因的全基因组鉴定和表达分析[J]. 生物技术通报, 2023, 39(1): 175-186. |
| [15] | 辛建攀, 李燕, 赵楚, 田如男. 镉胁迫下梭鱼草叶片转录组测序及苯丙烷代谢途径相关基因挖掘[J]. 生物技术通报, 2022, 38(6): 198-210. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||