








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 120-129.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0697
Previous Articles Next Articles
CHEN Kai-ling1( ), WU Tao1,2, XU Yi-qun1,2, GAO Jia1,2, ZHANG Mei-jun1,2, LI Xin1(
), WU Tao1,2, XU Yi-qun1,2, GAO Jia1,2, ZHANG Mei-jun1,2, LI Xin1( ), JIA Ju-qing1,2(
), JIA Ju-qing1,2( )
)
Received:2023-07-18
Online:2024-02-26
Published:2024-03-13
Contact:
LI Xin, JIA Ju-qing
E-mail:ckl1228@qq.com;lixin@sxau.edu.cn;jiajuqing@sxau.edu.cn
CHEN Kai-ling, WU Tao, XU Yi-qun, GAO Jia, ZHANG Mei-jun, LI Xin, JIA Ju-qing. Identification of SSR Loci and Development of Polymorphic Markers in Whole Genome of Oat[J]. Biotechnology Bulletin, 2024, 40(2): 120-129.
| 亚组 Subgroup | 染色体 Chromosome | 染色体大小 Size of chromosome/Mb | SSR数量 Number of SSRs | SSR密度 SSR density/(SSR·Mb-1) | SSR距离 SSR distance/(kb·SSR-1) |
|---|---|---|---|---|---|
| A | 1A | 551.55 | 46 585 | 84.46 | 11.84 |
| 2A | 462.81 | 41 810 | 90.34 | 11.07 | |
| 3A | 418.65 | 34 491 | 82.39 | 12.14 | |
| 4A | 468.67 | 41 615 | 88.79 | 11.26 | |
| 5A | 489.91 | 40 918 | 83.52 | 11.97 | |
| 6A | 456.82 | 40 619 | 88.92 | 11.25 | |
| 7A | 497.83 | 41 202 | 82.76 | 12.08 | |
| C | 1C | 459.16 | 30 835 | 67.15 | 14.89 |
| 2C | 574.16 | 38 944 | 67.83 | 14.74 | |
| 3C | 616.20 | 49 132 | 79.73 | 12.54 | |
| 4C | 701.82 | 46 713 | 66.56 | 15.02 | |
| 5C | 601.56 | 41 519 | 69.02 | 14.49 | |
| 6C | 610.20 | 45 292 | 74.22 | 13.47 | |
| 7C | 530.95 | 37 060 | 69.80 | 14.33 | |
| D | 1D | 473.71 | 36 983 | 78.07 | 12.80 |
| 2D | 532.98 | 41 375 | 77.63 | 12.88 | |
| 3D | 464.06 | 33 680 | 72.58 | 13.78 | |
| 4D | 451.84 | 37 661 | 83.35 | 12.00 | |
| 5D | 500.51 | 39 941 | 79.80 | 12.53 | |
| 6D | 293.84 | 22 043 | 75.02 | 13.33 | |
| 7D | 498.92 | 39 720 | 79.61 | 12.56 | |
| 合计Total | 10 656.15 | 828 138 | - | - | |
Table 1 SSR quantitative distribution characteristics
| 亚组 Subgroup | 染色体 Chromosome | 染色体大小 Size of chromosome/Mb | SSR数量 Number of SSRs | SSR密度 SSR density/(SSR·Mb-1) | SSR距离 SSR distance/(kb·SSR-1) |
|---|---|---|---|---|---|
| A | 1A | 551.55 | 46 585 | 84.46 | 11.84 |
| 2A | 462.81 | 41 810 | 90.34 | 11.07 | |
| 3A | 418.65 | 34 491 | 82.39 | 12.14 | |
| 4A | 468.67 | 41 615 | 88.79 | 11.26 | |
| 5A | 489.91 | 40 918 | 83.52 | 11.97 | |
| 6A | 456.82 | 40 619 | 88.92 | 11.25 | |
| 7A | 497.83 | 41 202 | 82.76 | 12.08 | |
| C | 1C | 459.16 | 30 835 | 67.15 | 14.89 |
| 2C | 574.16 | 38 944 | 67.83 | 14.74 | |
| 3C | 616.20 | 49 132 | 79.73 | 12.54 | |
| 4C | 701.82 | 46 713 | 66.56 | 15.02 | |
| 5C | 601.56 | 41 519 | 69.02 | 14.49 | |
| 6C | 610.20 | 45 292 | 74.22 | 13.47 | |
| 7C | 530.95 | 37 060 | 69.80 | 14.33 | |
| D | 1D | 473.71 | 36 983 | 78.07 | 12.80 |
| 2D | 532.98 | 41 375 | 77.63 | 12.88 | |
| 3D | 464.06 | 33 680 | 72.58 | 13.78 | |
| 4D | 451.84 | 37 661 | 83.35 | 12.00 | |
| 5D | 500.51 | 39 941 | 79.80 | 12.53 | |
| 6D | 293.84 | 22 043 | 75.02 | 13.33 | |
| 7D | 498.92 | 39 720 | 79.61 | 12.56 | |
| 合计Total | 10 656.15 | 828 138 | - | - | |
| 项目 Item | 平均值 Average | 标准差 Standard deviation | 染色体大小 Size of chromosome | SSR密度 SSR density | SSR距离 SSR distance | SSR数量 Number of SSRs |
|---|---|---|---|---|---|---|
| 染色体大小Size of chromosome | 507.44 | 85.19 | 1 | |||
| SSR密度SSR density | 78.16 | 7.38 | -0.38 | 1 | ||
| SSR距离SSR distance | 12.90 | 1.24 | 0.40 | -0.99** | 1 | |
| SSR数量Number of SSRs | 39 435.14 | 5 945.73 | 0.81** | 0.21 | -0.20 | 1 |
Table 2 Correlation analysis
| 项目 Item | 平均值 Average | 标准差 Standard deviation | 染色体大小 Size of chromosome | SSR密度 SSR density | SSR距离 SSR distance | SSR数量 Number of SSRs |
|---|---|---|---|---|---|---|
| 染色体大小Size of chromosome | 507.44 | 85.19 | 1 | |||
| SSR密度SSR density | 78.16 | 7.38 | -0.38 | 1 | ||
| SSR距离SSR distance | 12.90 | 1.24 | 0.40 | -0.99** | 1 | |
| SSR数量Number of SSRs | 39 435.14 | 5 945.73 | 0.81** | 0.21 | -0.20 | 1 |
| 基因组Genome | 亚组Subgroup | 染色体长度Chromosome length/Mb | SSR数目Number of SSR | SSR密度SSR density/(SSR·Mb-1) |
|---|---|---|---|---|
| Sang | A | 3 277.34 | 242 707 | 73.99 |
| C | 3 867.70 | 260 585 | 67.30 | |
| D | 3 125.40 | 223 135 | 71.32 | |
| 三分三 | A | 3 346.24 | 287 240 | 85.88 |
| C | 4 094.05 | 289 495 | 70.62 | |
| D | 3 215.86 | 251 403 | 78.01 |
Table 3 SSR distribution characteristics among subgroups
| 基因组Genome | 亚组Subgroup | 染色体长度Chromosome length/Mb | SSR数目Number of SSR | SSR密度SSR density/(SSR·Mb-1) |
|---|---|---|---|---|
| Sang | A | 3 277.34 | 242 707 | 73.99 |
| C | 3 867.70 | 260 585 | 67.30 | |
| D | 3 125.40 | 223 135 | 71.32 | |
| 三分三 | A | 3 346.24 | 287 240 | 85.88 |
| C | 4 094.05 | 289 495 | 70.62 | |
| D | 3 215.86 | 251 403 | 78.01 |
| 重复次数 Number of repeats | 重复类型Repeat type | 合计 Total | 占比 Proportion/% | |||||
|---|---|---|---|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 双核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | |||
| 5 | - | 0 | 164 814 | 8 794 | 1 467 | 2 281 | 177 356 | 21.42 |
| 6 | - | 131 050 | 50 279 | 2 990 | 427 | 970 | 185 716 | 22.43 |
| 7 | - | 56 606 | 20 222 | 1 181 | 179 | 526 | 78 714 | 9.50 |
| 8 | - | 28 448 | 10 737 | 583 | 77 | 316 | 40 161 | 4.85 |
| 9 | - | 14 827 | 6 875 | 361 | 49 | 207 | 22 319 | 2.70 |
| 10 | - | 7 751 | 4 958 | 223 | 26 | 165 | 13 123 | 1.58 |
| >10 | 247 167 | 40 281 | 21 936 | 816 | 96 | 453 | 310 749 | 37.52 |
| 合计Total | 247 167 | 278 963 | 279 821 | 14 948 | 2 321 | 4 918 | 828 138 | - |
| 占比Proportion /% | 29.85 | 33.69 | 33.79 | 1.81 | 0.28 | 0.59 | - | 100 |
Table 4 SSR repeat type, repeat number and proportion of whole genome of oat
| 重复次数 Number of repeats | 重复类型Repeat type | 合计 Total | 占比 Proportion/% | |||||
|---|---|---|---|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 双核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | |||
| 5 | - | 0 | 164 814 | 8 794 | 1 467 | 2 281 | 177 356 | 21.42 |
| 6 | - | 131 050 | 50 279 | 2 990 | 427 | 970 | 185 716 | 22.43 |
| 7 | - | 56 606 | 20 222 | 1 181 | 179 | 526 | 78 714 | 9.50 |
| 8 | - | 28 448 | 10 737 | 583 | 77 | 316 | 40 161 | 4.85 |
| 9 | - | 14 827 | 6 875 | 361 | 49 | 207 | 22 319 | 2.70 |
| 10 | - | 7 751 | 4 958 | 223 | 26 | 165 | 13 123 | 1.58 |
| >10 | 247 167 | 40 281 | 21 936 | 816 | 96 | 453 | 310 749 | 37.52 |
| 合计Total | 247 167 | 278 963 | 279 821 | 14 948 | 2 321 | 4 918 | 828 138 | - |
| 占比Proportion /% | 29.85 | 33.69 | 33.79 | 1.81 | 0.28 | 0.59 | - | 100 |
| 重复类型 Repeat type | 重复数目 Number of repeats | 优势重复基元 Dominant repeat primitives | SSR数目 Number of SSR | 占本重复类型百分比 Percentage of predominant motifs/% |
|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 4 | A/T | 145 340 | 58.80 |
| C/G | 101 827 | 41.20 | ||
| 双核苷酸 Dinucleotide | 12 | AG/CT | 137 111 | 49.15 |
| AT/AT | 69 342 | 24.86 | ||
| AC/GT | 67 856 | 24.32 | ||
| CG/CG | 4 654 | 1.67 | ||
| 三核苷酸 Trinucleotide | 60 | AAC/GTT | 68 530 | 24.49 |
| ATC/ATG | 57 361 | 20.50 | ||
| AAG/CTT | 45 805 | 16.37 | ||
| AGG/CCT | 28 650 | 10.24 | ||
| 其他 Others | 79 475 | 28.40 | ||
| 四核苷酸 Tetranucleotide | 238 | TTCA/TGAA | 3 968 | 26.55 |
| AAAT/ATTT | 2 286 | 15.29 | ||
| ATAG/CTAT | 1 753 | 11.73 | ||
| AAAG/CTTT | 1 003 | 6.71 | ||
| 其他 Others | 5 938 | 39.72 | ||
| 五核苷酸 Pentanucleotide | 390 | TAAGA/TCTTA | 910 | 39.21 |
| CCAAA/TTTGG | 186 | 8.01 | ||
| CGAGG/CCTCG | 176 | 7.58 | ||
| CGCCA/TGGC | 126 | 5.43 | ||
| 其他 Others | 923 | 39.77 | ||
| 六核苷酸 Hexanucleotide | 844 | CCTGGG/CCCAGG | 2 053 | 41.91 |
| AGGAGA/TCTCCT | 147 | 3.00 | ||
| TTGTTA/TAACAA | 142 | 2.90 | ||
| TTTAAA/TTTAAA | 130 | 2.65 | ||
| 其他 Others | 2 427 | 49.54 |
Table 5 Types and distribution of dominant motifs in each SSR repeat type of oat genome
| 重复类型 Repeat type | 重复数目 Number of repeats | 优势重复基元 Dominant repeat primitives | SSR数目 Number of SSR | 占本重复类型百分比 Percentage of predominant motifs/% |
|---|---|---|---|---|
| 单核苷酸 Mononucleotide | 4 | A/T | 145 340 | 58.80 |
| C/G | 101 827 | 41.20 | ||
| 双核苷酸 Dinucleotide | 12 | AG/CT | 137 111 | 49.15 |
| AT/AT | 69 342 | 24.86 | ||
| AC/GT | 67 856 | 24.32 | ||
| CG/CG | 4 654 | 1.67 | ||
| 三核苷酸 Trinucleotide | 60 | AAC/GTT | 68 530 | 24.49 |
| ATC/ATG | 57 361 | 20.50 | ||
| AAG/CTT | 45 805 | 16.37 | ||
| AGG/CCT | 28 650 | 10.24 | ||
| 其他 Others | 79 475 | 28.40 | ||
| 四核苷酸 Tetranucleotide | 238 | TTCA/TGAA | 3 968 | 26.55 |
| AAAT/ATTT | 2 286 | 15.29 | ||
| ATAG/CTAT | 1 753 | 11.73 | ||
| AAAG/CTTT | 1 003 | 6.71 | ||
| 其他 Others | 5 938 | 39.72 | ||
| 五核苷酸 Pentanucleotide | 390 | TAAGA/TCTTA | 910 | 39.21 |
| CCAAA/TTTGG | 186 | 8.01 | ||
| CGAGG/CCTCG | 176 | 7.58 | ||
| CGCCA/TGGC | 126 | 5.43 | ||
| 其他 Others | 923 | 39.77 | ||
| 六核苷酸 Hexanucleotide | 844 | CCTGGG/CCCAGG | 2 053 | 41.91 |
| AGGAGA/TCTCCT | 147 | 3.00 | ||
| TTGTTA/TAACAA | 142 | 2.90 | ||
| TTTAAA/TTTAAA | 130 | 2.65 | ||
| 其他 Others | 2 427 | 49.54 |
| 引物 Primer | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') | 退火温度Annealing temperature/℃ | 在4D染色体起始位置Starting position/bp | 扩增长度Amplification length/bp | 基元类型Primitive type | 重复次数Number of repeats |
|---|---|---|---|---|---|---|---|
| A-1 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 54 | 402 056 122 | 195 | TC | 27 |
| A-2 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 54 | 403 098 125 | 184 | GA | 12 |
| A-3 | CCCATAGTAGACCCTTGCCAT | ACGAGAGACTGAAGTGAAGCT | 58 | 404 060 755 | 296 | TTC | 9 |
| A-4 | TATACAATGGACCTCGGCAGA | ACTGCGTACACTGCTAGCTAT | 58 | 413 048 093 | 227 | TA | 30 |
| A-5 | ATTCCGGACTCTGCTTACGAT | TATCCTCAGGTATGCGTGGTT | 58 | 414 065 870 | 232 | TACA | 20 |
| A-6 | TGCATAGTGTTACCAGCGAAC | CGGTTTGAAAGATAGGGGTGG | 58 | 415 521 048 | 291 | TA | 26 |
| A-7 | CTGCTCACCACCACAATTCAA | TGAGCTATTCTACCACGTCCC | 58 | 416 396 796 | 292 | TA | 22 |
| A-8 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 58 | 417 151 310 | 243 | GA | 52 |
| A-9 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 59 | 418 130 363 | 204 | TC | 39 |
| A-10 | CACTGCCTGGTAAACTTCGAT | TGCTCATGTCTATTAACGTCCA | 57 | 420 675 833 | 289 | ATT | 11 |
| A-11 | CTCAGGTGCAGAGGAGAACTT | TGTTGTGTCTTGAGCCTGTTG | 58 | 421 639 959 | 275 | GTT | 7 |
| A-12 | AGACAGAGCGAGTACTTGGAA | ATGGATGTGTGCACGTTTCAT | 58 | 422 813 420 | 231 | GATG | 8 |
| A-13 | AGTCTTTGAGTCCGTGTCCAT | ATGTGGAGGAGCAATGAAAGC | 58 | 423 221 780 | 276 | TCC | 12 |
| A-14 | AGGAGATCAATCGCGGTACAT | TGTCACTATCCCTGTCACTGG | 58 | 424 040 249 | 284 | AAG | 13 |
| A-15 | TCTAGCTTGCTGGTCCTTGTT | ACTAATTGACAACACCAGCCC | 58 | 424 831 385 | 271 | GAT | 9 |
| A-16 | TGAAATCTGCCGGGGAAATTC | AGTAGTCAACACCGGGAAAGT | 58 | 424 944 085 | 200 | ATC | 5 |
| A-17 | GACTTCTAGGTCGCTGGTTC | ACAAGCGAAGGAAGAATCAC | 56 | 424 853 458 | 253 | AG | 19 |
| A-18 | GTCGTTGTAATCCCGATTGT | CTTCCTCCCTGAGTTCGATA | 57 | 424 714 810 | 391 | TA | 38 |
| A-19 | TTCAAACTATGGGTGCACTT | GATGTAAGACTGGCCTCCTG | 57 | 424 623 031 | 362 | TA | 26 |
| A-20 | AGCGTGTTATGTGGTGTTTG | CCGTCGCGTAGAATAAAAGT | 57 | 424 595 914 | 349 | AT | 27 |
| A-21 | CGATGTTGTGAGGGACAATA | ATCGACTGGACAAATGGAGT | 57 | 423 715 272 | 364 | TA | 30 |
| A-22 | CTCTGACAGTCCAACTGCAA | AGTCGTCGTATGTTGTGTGG | 57 | 423 425 714 | 333 | CAA | 10 |
| A-23 | TTGGTGGTCTCTTGCTGTTA | TGGGCCAAAGAGAATTTTAG | 57 | 423 121 188 | 307 | TA | 49 |
| A-24 | TGATCCACCTCCAGGACTAT | CCATCTCTAGGCCTTCACTG | 57 | 422 863 900 | 394 | GA | 26 |
| A-25 | TATCATCGAGTTCCGACTCA | TAGTGTCCTGCATGGTTGTC | 57 | 422 092 205 | 344 | TTC | 10 |
| A-26 | AGTGGTGGGAGTGAGGTGT | CACCTCCAAGCCCTAAACTA | 57 | 422 030 145 | 260 | GA | 81 |
| A-27 | GGAAAACACCGAGAGAGAAA | TGTCCTCAAGAACAAAACGA | 57 | 421 993 919 | 378 | ATC | 8 |
| A-28 | ACTAAAGGTTGCTCCGTTTG | TGAGCTTGCACATCATTTCT | 57 | 421 827 958 | 331 | AGA | 6 |
| A-29 | CTCCCTCCGTTTTGAAAATA | CCATCAACGGAACATGTAAA | 57 | 420 311 577 | 366 | AT | 30 |
| A-30 | TGTGGTGAATATGCCTCTTG | AAAGGAAAAGGGAGAAAGGA | 57 | 420 091 310 | 369 | AG | 30 |
| A-31 | GGAGTCGTTCTTGATGTTGA | ATAATCCAATCCAACCCAAA | 57 | 419 525 753 | 365 | CGA | 9 |
| A-32 | AAACCTGACTGGACCTTCTG | GATGGCAATCCAACAGTTTT | 57 | 418 055 976 | 265 | TA | 27 |
| A-33 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 823 067 | 398 | AG | 43 |
| A-34 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 740 723 | 393 | AT | 36 |
| A-35 | TGGATCACTCATACGGTCAG | GAGGGGTCATTGTCTACAGG | 57 | 416 846 114 | 391 | GA | 46 |
| A-36 | GGTAAACGCAAAGCTGAAAT | AAAGGGCATGGTATCCATTA | 57 | 416 404 269 | 324 | GA | 6 |
| A-37 | TCGAAAGGGAATGTTCTAGC | TTGCTCCTACTGCCACATTA | 57 | 415 943 775 | 312 | CT | 20 |
| A-38 | TTTGGGAGGACCTCTAGTTG | TACCCTTGAAGACCGAAAAG | 57 | 415 647 138 | 370 | AAC | 5 |
| A-39 | ATGCAACAAATGATGAATGC | TAGTGCGAAAACCATCTTCA | 57 | 414 940 663 | 394 | CT | 16 |
| A-40 | CTAACTGAGCGTCGCATAAC | CTCAAGGGATGGGGATAAG | 57 | 414 641 081 | 351 | AT | 30 |
| A-41 | TGTTCATGTATCCCGCATAG | TTCCAGACGCCTAGGTTTAG | 57 | 414 586 340 | 359 | AT | 31 |
| A-42 | TGAAAGATAAGGGGTGGATG | AAGTGCACCCATAGTTTGAA | 57 | 414 263 854 | 378 | TA | 13 |
| A-43 | GCCATTATTTGTGCATGAAG | ATGTGGGGTGCAAAATAACT | 57 | 413 518 395 | 355 | TA | 34 |
| A-44 | AGTGCATCGAACTCTCTTCC | GCTGCGTACGTACATCTCAA | 57 | 413 420 890 | 337 | TC | 46 |
| A-45 | TGATGAATCGAGAAGACACG | ATTCGCAACATGAAAAACAA | 57 | 413 194 121 | 377 | ATG | 13 |
| A-46 | CAAGATGCGGATGATGATTA | GAACCTTCGATACCACAACC | 57 | 412 897 229 | 365 | TC | 6 |
| A-47 | ATGACTTTCCGCAATCTCTC | AAGGTTTGACACGATGACCT | 57 | 411 869 015 | 316 | TTC | 24 |
| A-48 | CCCAAATAGACCAAGGAATG | ATACTGAGCAGGCAAAGGAC | 57 | 411 852 406 | 370 | AT | 40 |
| A-49 | AAAAGCCCTGACTAAGAGCA | TTCCTTTGGTCTCAAAATGG | 57 | 409 342 863 | 396 | TA | 40 |
| A-50 | GGGAAAGATCAATGGTTCAG | TTCCAAAAGCTGACACGTTA | 57 | 408 678 582 | 259 | TA | 33 |
| A-51 | GGTGGTTAGATGGGATCAGA | ACTCTGGGTGTTGAATAGGG | 57 | 406 013 380 | 347 | TC | 31 |
| A-52 | TAGAATTCCTCCCACATGGT | CGATCCTTGCAGTTAGGAGT | 57 | 405 122 840 | 239 | GA | 12 |
Table 6 Information of SSR primers developed for oats polymorphism
| 引物 Primer | 正向引物 Forward primer(5'-3') | 反向引物 Reverse primer(5'-3') | 退火温度Annealing temperature/℃ | 在4D染色体起始位置Starting position/bp | 扩增长度Amplification length/bp | 基元类型Primitive type | 重复次数Number of repeats |
|---|---|---|---|---|---|---|---|
| A-1 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 54 | 402 056 122 | 195 | TC | 27 |
| A-2 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 54 | 403 098 125 | 184 | GA | 12 |
| A-3 | CCCATAGTAGACCCTTGCCAT | ACGAGAGACTGAAGTGAAGCT | 58 | 404 060 755 | 296 | TTC | 9 |
| A-4 | TATACAATGGACCTCGGCAGA | ACTGCGTACACTGCTAGCTAT | 58 | 413 048 093 | 227 | TA | 30 |
| A-5 | ATTCCGGACTCTGCTTACGAT | TATCCTCAGGTATGCGTGGTT | 58 | 414 065 870 | 232 | TACA | 20 |
| A-6 | TGCATAGTGTTACCAGCGAAC | CGGTTTGAAAGATAGGGGTGG | 58 | 415 521 048 | 291 | TA | 26 |
| A-7 | CTGCTCACCACCACAATTCAA | TGAGCTATTCTACCACGTCCC | 58 | 416 396 796 | 292 | TA | 22 |
| A-8 | TCCTTAGGCCGTGAAGAAGTT | TGCTCTATGTACTGATCCGTGA | 58 | 417 151 310 | 243 | GA | 52 |
| A-9 | ATGGTAAACGGGTCATCGAGT | AAGTATAGGCCGACGATCCAG | 59 | 418 130 363 | 204 | TC | 39 |
| A-10 | CACTGCCTGGTAAACTTCGAT | TGCTCATGTCTATTAACGTCCA | 57 | 420 675 833 | 289 | ATT | 11 |
| A-11 | CTCAGGTGCAGAGGAGAACTT | TGTTGTGTCTTGAGCCTGTTG | 58 | 421 639 959 | 275 | GTT | 7 |
| A-12 | AGACAGAGCGAGTACTTGGAA | ATGGATGTGTGCACGTTTCAT | 58 | 422 813 420 | 231 | GATG | 8 |
| A-13 | AGTCTTTGAGTCCGTGTCCAT | ATGTGGAGGAGCAATGAAAGC | 58 | 423 221 780 | 276 | TCC | 12 |
| A-14 | AGGAGATCAATCGCGGTACAT | TGTCACTATCCCTGTCACTGG | 58 | 424 040 249 | 284 | AAG | 13 |
| A-15 | TCTAGCTTGCTGGTCCTTGTT | ACTAATTGACAACACCAGCCC | 58 | 424 831 385 | 271 | GAT | 9 |
| A-16 | TGAAATCTGCCGGGGAAATTC | AGTAGTCAACACCGGGAAAGT | 58 | 424 944 085 | 200 | ATC | 5 |
| A-17 | GACTTCTAGGTCGCTGGTTC | ACAAGCGAAGGAAGAATCAC | 56 | 424 853 458 | 253 | AG | 19 |
| A-18 | GTCGTTGTAATCCCGATTGT | CTTCCTCCCTGAGTTCGATA | 57 | 424 714 810 | 391 | TA | 38 |
| A-19 | TTCAAACTATGGGTGCACTT | GATGTAAGACTGGCCTCCTG | 57 | 424 623 031 | 362 | TA | 26 |
| A-20 | AGCGTGTTATGTGGTGTTTG | CCGTCGCGTAGAATAAAAGT | 57 | 424 595 914 | 349 | AT | 27 |
| A-21 | CGATGTTGTGAGGGACAATA | ATCGACTGGACAAATGGAGT | 57 | 423 715 272 | 364 | TA | 30 |
| A-22 | CTCTGACAGTCCAACTGCAA | AGTCGTCGTATGTTGTGTGG | 57 | 423 425 714 | 333 | CAA | 10 |
| A-23 | TTGGTGGTCTCTTGCTGTTA | TGGGCCAAAGAGAATTTTAG | 57 | 423 121 188 | 307 | TA | 49 |
| A-24 | TGATCCACCTCCAGGACTAT | CCATCTCTAGGCCTTCACTG | 57 | 422 863 900 | 394 | GA | 26 |
| A-25 | TATCATCGAGTTCCGACTCA | TAGTGTCCTGCATGGTTGTC | 57 | 422 092 205 | 344 | TTC | 10 |
| A-26 | AGTGGTGGGAGTGAGGTGT | CACCTCCAAGCCCTAAACTA | 57 | 422 030 145 | 260 | GA | 81 |
| A-27 | GGAAAACACCGAGAGAGAAA | TGTCCTCAAGAACAAAACGA | 57 | 421 993 919 | 378 | ATC | 8 |
| A-28 | ACTAAAGGTTGCTCCGTTTG | TGAGCTTGCACATCATTTCT | 57 | 421 827 958 | 331 | AGA | 6 |
| A-29 | CTCCCTCCGTTTTGAAAATA | CCATCAACGGAACATGTAAA | 57 | 420 311 577 | 366 | AT | 30 |
| A-30 | TGTGGTGAATATGCCTCTTG | AAAGGAAAAGGGAGAAAGGA | 57 | 420 091 310 | 369 | AG | 30 |
| A-31 | GGAGTCGTTCTTGATGTTGA | ATAATCCAATCCAACCCAAA | 57 | 419 525 753 | 365 | CGA | 9 |
| A-32 | AAACCTGACTGGACCTTCTG | GATGGCAATCCAACAGTTTT | 57 | 418 055 976 | 265 | TA | 27 |
| A-33 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 823 067 | 398 | AG | 43 |
| A-34 | ACTTTGGTTGCTCTCTTTGC | GAAACCTCCGTATTGTCGAT | 57 | 417 740 723 | 393 | AT | 36 |
| A-35 | TGGATCACTCATACGGTCAG | GAGGGGTCATTGTCTACAGG | 57 | 416 846 114 | 391 | GA | 46 |
| A-36 | GGTAAACGCAAAGCTGAAAT | AAAGGGCATGGTATCCATTA | 57 | 416 404 269 | 324 | GA | 6 |
| A-37 | TCGAAAGGGAATGTTCTAGC | TTGCTCCTACTGCCACATTA | 57 | 415 943 775 | 312 | CT | 20 |
| A-38 | TTTGGGAGGACCTCTAGTTG | TACCCTTGAAGACCGAAAAG | 57 | 415 647 138 | 370 | AAC | 5 |
| A-39 | ATGCAACAAATGATGAATGC | TAGTGCGAAAACCATCTTCA | 57 | 414 940 663 | 394 | CT | 16 |
| A-40 | CTAACTGAGCGTCGCATAAC | CTCAAGGGATGGGGATAAG | 57 | 414 641 081 | 351 | AT | 30 |
| A-41 | TGTTCATGTATCCCGCATAG | TTCCAGACGCCTAGGTTTAG | 57 | 414 586 340 | 359 | AT | 31 |
| A-42 | TGAAAGATAAGGGGTGGATG | AAGTGCACCCATAGTTTGAA | 57 | 414 263 854 | 378 | TA | 13 |
| A-43 | GCCATTATTTGTGCATGAAG | ATGTGGGGTGCAAAATAACT | 57 | 413 518 395 | 355 | TA | 34 |
| A-44 | AGTGCATCGAACTCTCTTCC | GCTGCGTACGTACATCTCAA | 57 | 413 420 890 | 337 | TC | 46 |
| A-45 | TGATGAATCGAGAAGACACG | ATTCGCAACATGAAAAACAA | 57 | 413 194 121 | 377 | ATG | 13 |
| A-46 | CAAGATGCGGATGATGATTA | GAACCTTCGATACCACAACC | 57 | 412 897 229 | 365 | TC | 6 |
| A-47 | ATGACTTTCCGCAATCTCTC | AAGGTTTGACACGATGACCT | 57 | 411 869 015 | 316 | TTC | 24 |
| A-48 | CCCAAATAGACCAAGGAATG | ATACTGAGCAGGCAAAGGAC | 57 | 411 852 406 | 370 | AT | 40 |
| A-49 | AAAAGCCCTGACTAAGAGCA | TTCCTTTGGTCTCAAAATGG | 57 | 409 342 863 | 396 | TA | 40 |
| A-50 | GGGAAAGATCAATGGTTCAG | TTCCAAAAGCTGACACGTTA | 57 | 408 678 582 | 259 | TA | 33 |
| A-51 | GGTGGTTAGATGGGATCAGA | ACTCTGGGTGTTGAATAGGG | 57 | 406 013 380 | 347 | TC | 31 |
| A-52 | TAGAATTCCTCCCACATGGT | CGATCCTTGCAGTTAGGAGT | 57 | 405 122 840 | 239 | GA | 12 |
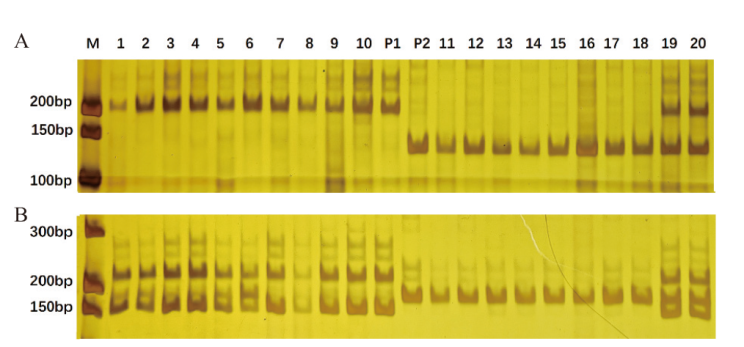
Fig. 2 Amplification results of primers A-1(A)and A-2(B)in experimental materials M: DNA marker; P1: Banner; P2: Baiyan 2; 1-20: F2 20 individual plants
| [1] | 吴斌, 郑殿升, 严威凯, 等. 燕麦分子育种研究进展[J]. 植物遗传资源学报, 2019, 20(3): 485-495. |
| Wu B, Zheng DS, Yan KW, et al. Advances in molecular breeding of oats[J]. Journal of Plant Genetic Resources, 2019, 20(3): 485-495. | |
| [2] | Rasane P, Jha A, Sabikhi L, et al. Nutritional advantages of oats and opportunities for its processing as value added foods-areview[J]. J Food Sci Technol, 2015, 52(2): 662-675. |
| [3] |
潘莹, 程时锋. 燕麦基因组学研究进展[J]. 植物遗传资源学报, 2021, 22(2): 304-308.
doi: 10.13430/j.cnki.jpgr.20200910001 |
| Pan Y, Cheng SF. Research progress on oat genomics study[J]. J Plant Genet Res, 2021, 22(2): 304-308. | |
| [4] |
Kamal N, Tsardakas Renhuldt N, Bentzer J, et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop[J]. Nature, 2022, 606(7912): 113-119.
doi: 10.1038/s41586-022-04732-y |
| [5] |
Peng YY, Yan HH, Guo LC, et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat[J]. Nat Genet, 2022, 54(8): 1248-1258.
doi: 10.1038/s41588-022-01127-7 |
| [6] | Wu JZ, Zhao Q, Wu GW, et al. Development of novel SSR markers for flax(Linum usitatissimum L.)using reduced-representation genome sequencing[J]. Front Plant Sci, 2017, 7: 2018. |
| [7] |
Guo R, Landis JB, Moore MJ, et al. Development and application of transcriptome-derived microsatellites in Actinidia eriantha(Actinidiaceae)[J]. Front Plant Sci, 2017, 8: 1383.
doi: 10.3389/fpls.2017.01383 URL |
| [8] |
Malausa T, Gilles A, Meglecz E, et al. High-throughput microsatellite isolation through 454 GS-FLX titanium pyrosequencing of enriched DNA libraries[J]. Mol Ecol Resour, 2011, 11(4): 638-644.
doi: 10.1111/j.1755-0998.2011.02992.x pmid: 21676194 |
| [9] |
Sowa S, Paczos-Grzęda E. Identification of molecular markers for the PC39 gene conferring resistance to crown rust in oat[J]. Theor Appl Genet, 2020, 133(4): 1081-1094.
doi: 10.1007/s00122-020-03533-z pmid: 31927607 |
| [10] |
Zhao J, Kebede AZ, Menzies JG, et al. Chromosomal location of the crown rust resistance gene Pc98 in cultivated oat(Avena sativa L.)[J]. Theor Appl Genet, 2020, 133(4): 1109-1122.
doi: 10.1007/s00122-020-03535-x pmid: 31938813 |
| [11] |
Yan H, Yu K, Xu Y, et al. Position validation of the dwarfing gene dw6 in oat(Avena sativa L.)and its correlated effects on agronomic traits[J]. Front Plant Sci, 2021, 12: 668847.
doi: 10.3389/fpls.2021.668847 URL |
| [12] |
吴斌, 张茜, 宋高原, 等. 裸燕麦SSR标记连锁群图谱的构建及β-葡聚糖含量QTL的定位[J]. 中国农业科学, 2014, 47(6): 1208-1215.
doi: 10.3864/j.issn.0578-1752.2014.06.017 |
| Wu B, Zhang Q, Song GY, et al. Construction of SSR genetic linkage map and analysis of QTLs related to β-glucan Content of naked oat(Avena nuda L.)[J]. Sci Agric Sin, 2014, 47(6): 1208-1215. | |
| [13] |
Isabel LP, Park JR, Lee GS, et al. Development of EST-SSR markers and analysis of genetic relationship it's resources in hexaploid oats[J]. J Crop Sci Biotechnol, 2019, 22(3): 243-251.
doi: 10.1007/s12892-019-0158-0 |
| [14] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [15] |
Ubert IP, Zimmer CM, Pellizzaro K, et al. Genetics and molecular mapping of the naked grains in hexaploid oat[J]. Euphytica, 2017, 213(2): 41.
doi: 10.1007/s10681-017-1836-1 |
| [16] |
Zhou Y, Zhao XB, Li YW, et al. Triticum population sequencing provides insights into wheat adaptation[J]. Nat Genet, 2020, 52(12): 1412-1422.
doi: 10.1038/s41588-020-00722-w pmid: 33106631 |
| [17] | 原志敏. 玉米全基因组SSRs分子标记开发与特征分析[D]. 雅安: 四川农业大学, 2013. |
| Yuan ZM. Development and characterization of SSRs molecular markers in maize genome[D]. Ya'an: Sichuan Agricultural University, 2013. | |
| [18] | 马名川, 刘龙龙, 刘璋, 等. 苦荞全基因组SSR位点特征分析与分子标记开发[J]. 作物杂志, 2021(1): 38-46. |
| Ma MC, Liu LL, Liu Z, et al. Analysis of SSR loci in whole genome and development of molecular markers in tartary buckwheat[J]. Crops, 2021(1): 38-46. | |
| [19] | 张晗, 王雪梅, 王东建, 等. 谷子基因组SSR信息分析和标记开发[J]. 分子植物育种, 2013, 11(1): 30-36. |
| Zhang H, Wang XM, Wang DJ, et al. Survey of SSRs in foxtail millet genome and development of SSR markers[J]. Mol Plant Breed, 2013, 11(1): 30-36. | |
| [20] | 仇静静. 野生花生全基因组SSR标记的开发与应用[D]. 济南: 山东师范大学, 2018. |
| QIU JJ. Development and application of SSR markers in wild peanut genome[D]. Jinan: Shandong Normal University, 2018. | |
| [21] |
Cavagnaro PF, Senalik DA, Yang LM, et al. Genome-wide characterization of simple sequence repeats in cucumber(Cucumis sativus L.)[J]. BMC Genomics, 2010, 11: 569.
doi: 10.1186/1471-2164-11-569 pmid: 20950470 |
| [22] |
关玲, 章镇, 王新卫, 等. 苹果基因组SSR位点分析与应用[J]. 中国农业科学, 2011, 44(21): 4415-4428.
doi: 10.3864/j.issn.0578-1752.2011.21.010 |
|
Guan L, Zhang Z, Wang XW, et al. Evaluation and application of the SSR loci in apple genome[J]. Sci Agric Sin, 2011, 44(21): 4415-4428.
doi: 10.3864/j.issn.0578-1752.2011.21.010 |
|
| [23] |
李乔乔, 王宇晴, 刘蕊, 等. 甜菜全基因组SSR引物的筛选与评价[J]. 中国农学通报, 2022, 38(12): 95-99.
doi: 10.11924/j.issn.1000-6850.casb2021-0726 |
|
Li QQ, Wang YQ, Liu R, et al. The whole genome SSR primers of sugar beet: screening and evaluation[J]. Chin Agric Sci Bull, 2022, 38(12): 95-99.
doi: 10.11924/j.issn.1000-6850.casb2021-0726 |
|
| [24] |
Han B, Wang CB, Tang ZH, et al. Genome-wide analysis of microsatellite markers based on sequenced database in chinese spring wheat(Triticum aestivum L.)[J]. PLoS One, 2015, 10(11): e0141540.
doi: 10.1371/journal.pone.0141540 URL |
| [25] |
Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes[J]. Nat Genet, 2002, 30(2): 194-200.
doi: 10.1038/ng822 pmid: 11799393 |
| [26] |
张霞, 于卓, 金兴红, 等. 马铃薯SSR引物的开发、特征分析及在彩色马铃薯材料中的扩增研究[J]. 作物学报, 2022, 48(4): 920-929.
doi: 10.3724/SP.J.1006.2022.14065 |
|
Zhang X, Yu Z, Jin XH, et al. Development and characterization analysis of potato SSR primers and the amplification research in colored potato materials[J]. Acta Agron Sin, 2022, 48(4): 920-929.
doi: 10.3724/SP.J.1006.2022.14065 |
|
| [27] | 杜磊, 蒙秋伊, 尚昆, 等. 薏苡基因组SSR标记开发与应用[J]. 分子植物育种, 2022, 20(3): 887-894. |
| Du L, Meng QY, Shang K, et al. Development and application of genomic SSR markers in coix lachryma-jobi genome[J]. Mol Plant Breed, 2022, 20(3): 887-894. | |
| [28] |
姚嘉瑜, 张立武, 赵捷, 等. 黄麻全基因组SSR鉴定与特征分析[J]. 作物学报, 2019, 45(1): 10-17.
doi: 10.3724/SP.J.1006.2019.84072 |
|
Yao JY, Zhang LW, Zhao J, et al. Evaluation and characteristic analysis of SSRs from the whole genome of jute(Corchorus capsularis)[J]. Acta Agron Sin, 2019, 45(1): 10-17.
doi: 10.3724/SP.J.1006.2019.84072 URL |
|
| [29] |
童治军, 焦芳婵, 肖炳光. 普通烟草及其祖先种基因组SSR位点分析[J]. 中国农业科学, 2015, 48(11): 2108-2117.
doi: 10.3864/j.issn.0578-1752.2015.11.003 |
| Tong ZJ, Jiao FC, Xiao BG. Analysis of SSR loci in nicotina tabacum genome and its two ancestral species genome[J]. Sci Agric Sin, 2015, 48(11): 2108-2117. | |
| [30] |
叶卫军, 陈圣男, 杨勇, 等. 绿豆SSR标记的开发及遗传多样性分析[J]. 作物学报, 2019, 45(8): 1176-1188.
doi: 10.3724/SP.J.1006.2019.84155 |
| Ye WJ, Chen SN, Yang Y, et al. Development of SSR markers and genetic diversity analysis in mung bean[J]. Acta Agron Sin, 2019, 45(8): 1176-1188. | |
| [31] |
Oliveira EJ, Pádua JG, Zucchi MI, et al. Origin, evolution and genome distribution of microsatellites[J]. Genet Mol Biol, 2006, 29(2): 294-307.
doi: 10.1590/S1415-47572006000200018 URL |
| [32] |
Luo MC, Gu YQ, Puiu D, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii[J]. Nature, 2017, 551(7681): 498-502.
doi: 10.1038/nature24486 URL |
| [1] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [2] | YU Shi-zhou, CAO Ling-gai, WANG Shi-ze, LIU Yong, BIAN Wen-jie, REN Xue-liang. Development Core SNP Markers for Tobacco Germplasm Genotyping [J]. Biotechnology Bulletin, 2023, 39(3): 89-100. |
| [3] | AN Miao, WANG Tong-tong, FU Yi-ting, XIA Jun-jun, PENG Suo-tang, DUAN Yong-hong. Genetic Diversity Analysis and Molecular Identity Card Construction by SSR Markers of 52 Solanum tuberosum L. Varieties(Lines) [J]. Biotechnology Bulletin, 2023, 39(12): 136-147. |
| [4] | ZHANG Ting-huan, ZHANG Li-juan, CHEN Si-qing, GUO Zong-yi. Effects of the Polymorphism of the Seed Sequence in Porcine miR-378 on Its Function and Carcass Traits [J]. Biotechnology Bulletin, 2021, 37(6): 154-162. |
| [5] | GUO Li-li, LI Yu-ying, GUO Da-long, HOU Xiao-gai. Research Progress on High-density Genetic Linkage Map Construction of Important Ornamental Plants:a Review [J]. Biotechnology Bulletin, 2021, 37(1): 246-254. |
| [6] | CHEN Yi-dan, ZHANG Yu, YANG Jie, ZHANG Qin, JIANG Li. Exploration of Key Functional Genes Affecting Milk Production Traits in Dairy Cattle Based on RNA-seq [J]. Biotechnology Bulletin, 2020, 36(9): 244-252. |
| [7] | ZHANG De-rong, MA Xiao-xia, LI Yu-fei, ZHAO Yong-qing, HUO Sheng-dong, MA Zhong-ren, BAI Jia-lin. Research Progress and Prospects of Adiponectin and Its Receptor in Mammal [J]. Biotechnology Bulletin, 2020, 36(6): 236-244. |
| [8] | YU Jun-jian, CHI Mei-li, JIA Yong-yi, LIU Shi-li, ZHU Jun-quan, GU Zhi-min. Tetra-primer Amplification Refractory Mutation System PCR and Its Application in Fauna and Flora Genetics and Breeding Research [J]. Biotechnology Bulletin, 2020, 36(5): 32-38. |
| [9] | LI Xiao-kai, FAN Yi-xing, QIAO Xian, ZHANG Lei, WANG Feng-hong, WANG Zhi-ying, WANG Rui-jun, ZHANG Yan-jun, LIU Zhi-hong, WANG Zhi-xin, HE Li-bing, LI Jin-quan, SU Rui, ZHANG Jia-xin. Research Progress of Goat Genome and Genetic Variation Map [J]. Biotechnology Bulletin, 2020, 36(4): 175-184. |
| [10] | CHANG Yong-fang, BAO Peng-jia, CHU Min, WU Xiao-yun, LIANG Chun-nian, YAN Ping. Research Progress on the Regulation of LncRNA in the Development of Mammalian Hair Follicle [J]. Biotechnology Bulletin, 2019, 35(8): 205-212. |
| [11] | LI Biao, ZHANG Rui-ying, WANG Xiao-qi, ZHANG Cun-fang, DUAN Zi-yuan. Microsatellite Polymorphism and Its Correlation Analysis with Body Size Traits of Tan Sheep [J]. Biotechnology Bulletin, 2019, 35(6): 131-137. |
| [12] | HUANG Long, WU Ben-li, HE Ji-xiang, CHEN Jing, SONG Guang-tong, WANG Xiang, ZHANG Ye, WU Song. SNP Identification of MyoD1 Gene and Its Correlation with Growth Traits in Pelodiscus sinensis [J]. Biotechnology Bulletin, 2019, 35(4): 76-81. |
| [13] | GUO Meng-meng, ZHOU Yan-qing, DUAN Hong-ying, YANG Ke, SHAO Lu-ying. Development of SNP Marker Based on the Rehmannia glutinosa Transcriptome Database and Construction of DNA Fingerprint in Rehmannia [J]. Biotechnology Bulletin, 2019, 35(11): 224-230. |
| [14] | WANG Yan-xin, LIAO Yuan-yuan, A Yimuguli, QI Ao-qiong, LI Hai-jian, XU Hong-wei, YANG Ju-tian, CAI Yong. Correlation Analysis Between LHX3 Gene Polymorphism and Growth Traits in Three Sheep Breeds [J]. Biotechnology Bulletin, 2019, 35(10): 162-168. |
| [15] | LI Sha-sha, ZHANG Liang, SHI Gui-yang. Screening of Microsatellite Markers Associated with Furfural Tolerance of Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2018, 34(8): 123-129. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||