








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (4): 8-17.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0927
Previous Articles Next Articles
LUO Wei1( ), MU Qiong2, SHU Jian-hong2, WU Jia-hai1,3(
), MU Qiong2, SHU Jian-hong2, WU Jia-hai1,3( ), WANG Xiao-li2(
), WANG Xiao-li2( )
)
Received:2020-07-24
Online:2021-04-26
Published:2021-05-13
Contact:
WU Jia-hai,WANG Xiao-li
E-mail:1291928810@qq.com;848266168@qq.com;wangxiaolizhenyuan@126.com
LUO Wei, MU Qiong, SHU Jian-hong, WU Jia-hai, WANG Xiao-li. Expression,Protein Interactions and Biological Function Analysis of FaFT in Festuca arundinacea[J]. Biotechnology Bulletin, 2021, 37(4): 8-17.
| 引物Primer | 序列Sequence(5'-3') | 用途Purpose |
|---|---|---|
| FaFT-F1 | GCGGGAACCGTTGTCTCTAT | 基因组扩增Genome amplification |
| FaFT-R1 | GTCCGGTCACGACCTTCAAT | |
| FaFT-F2 | GGATCCTTTGGCATTGAGCC | 实时荧光定量PCR Real-time fluorescence quantitative PCR |
| FaFT-R2 | CCGGTCACGACCTTCAATCT | |
| FaGAPDH-F | ACCCCTTCATCACCACCGACTAC | 内参基因Reference gene |
| FaGAPDH-R | TCCTTCTCGTTGACACCCATGAC | |
| FaFT-BD-F | GCATATGGCCATGGAGGCCGAATTC | 酵母双杂交Yeast two-hybrid |
| FaFT-BD-R | GCGGCCGCTGCAGGTCGACGGATCC | |
| FaFD-AD-F | TATGGCCATGGAGGCCAGTGAATTC | |
| FaFD-AD-R | TCTGCAGCTCGAGCTCGATGGATCC | |
| FaFT-C-F | GCCTGGCGCGCCACTAGTGGATCCA | 双分子荧光互补Bimolecular fluorescence complementation |
| FaFT-C-R | CATCCCGGGAGCGGTACCCTCGAG | |
| Primer F | GGGGTACCATGTCGCGGGGCAGGGAT | 过表达载体构建Overexpression vector construction |
| Primer R | GCTCTAGATTAGCTTGTTCCTTGGAA |
Table 1 Primer sequence
| 引物Primer | 序列Sequence(5'-3') | 用途Purpose |
|---|---|---|
| FaFT-F1 | GCGGGAACCGTTGTCTCTAT | 基因组扩增Genome amplification |
| FaFT-R1 | GTCCGGTCACGACCTTCAAT | |
| FaFT-F2 | GGATCCTTTGGCATTGAGCC | 实时荧光定量PCR Real-time fluorescence quantitative PCR |
| FaFT-R2 | CCGGTCACGACCTTCAATCT | |
| FaGAPDH-F | ACCCCTTCATCACCACCGACTAC | 内参基因Reference gene |
| FaGAPDH-R | TCCTTCTCGTTGACACCCATGAC | |
| FaFT-BD-F | GCATATGGCCATGGAGGCCGAATTC | 酵母双杂交Yeast two-hybrid |
| FaFT-BD-R | GCGGCCGCTGCAGGTCGACGGATCC | |
| FaFD-AD-F | TATGGCCATGGAGGCCAGTGAATTC | |
| FaFD-AD-R | TCTGCAGCTCGAGCTCGATGGATCC | |
| FaFT-C-F | GCCTGGCGCGCCACTAGTGGATCCA | 双分子荧光互补Bimolecular fluorescence complementation |
| FaFT-C-R | CATCCCGGGAGCGGTACCCTCGAG | |
| Primer F | GGGGTACCATGTCGCGGGGCAGGGAT | 过表达载体构建Overexpression vector construction |
| Primer R | GCTCTAGATTAGCTTGTTCCTTGGAA |
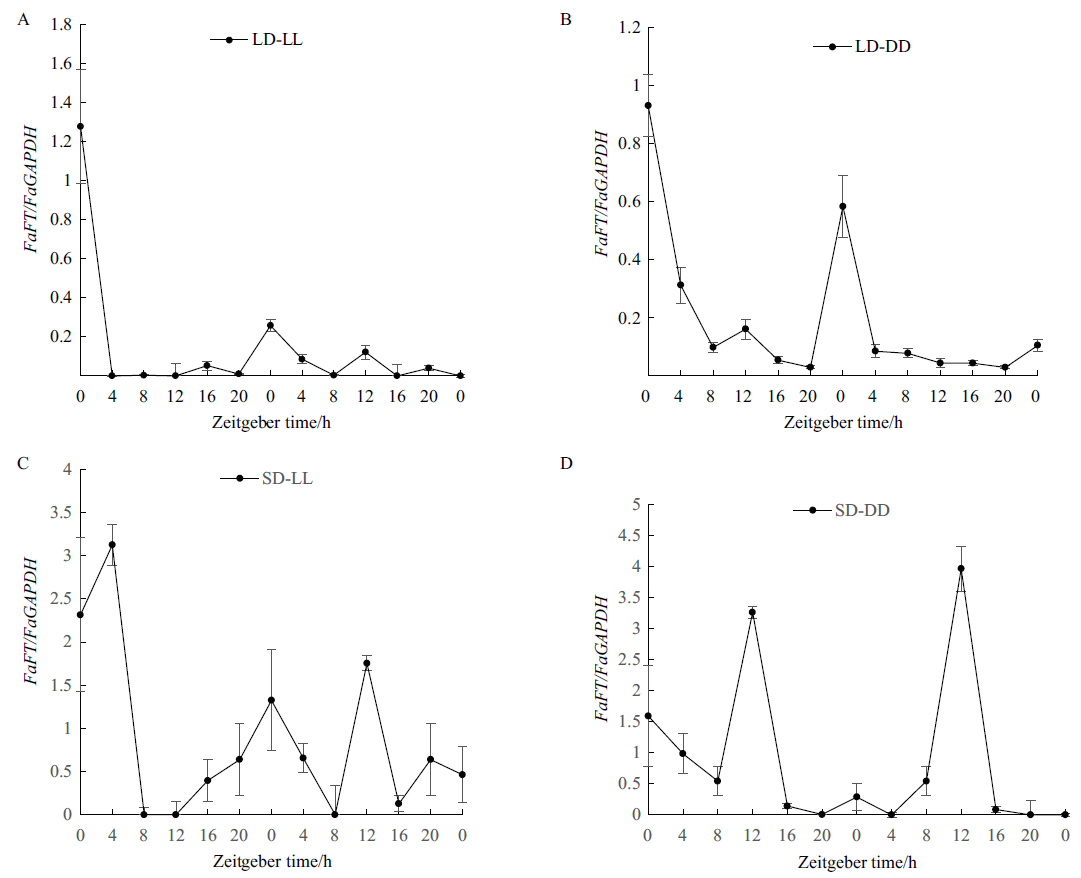
Fig.3 Expression level of FaFT gene of tall fescue from sunlight to continuous light (A) and continuous darkness (B) based on long day, continuous light (C) and continuous darkness (D) based on short day
| [1] |
Qin ZR, Wu JJ, Geng SF, et al. Regulation of FT splicing by an endogenous cue in temperate grasses[J]. Nat Commun, 2017. DOI: 10. 1038/ncomms14320.
doi: 10. 1038/ncomms14320 |
| [2] | 吕波. 植物开花基因FT的遗传转化及其参与开花调控的研究[D]. 泰安:山东农业大学, 2014. |
| Lv B. Genetic transformation of plant flowering gene FT and its involvement in flowering control[D]. Tai’an:Shandong Agriculture University, 2014. | |
| [3] |
Andres F, Kinoshita A, et al. The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana[J]. BMC Plant Biol, 2020,20(1):53.
doi: 10.1186/s12870-020-2266-0 URL |
| [4] |
Wigge PA, Kim MC, Jaeger KE, et al. Integration of spatial and temporal information during floral induction in Arabidopsis[J]. Science, 2005,309(5737):1056-1059.
doi: 10.1126/science.1114358 URL |
| [5] |
Böhlenius H, Huang T, Charbonnel-Campaa L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees[J]. Science, 2006,312(5776):1040-1043.
doi: 10.1126/science.1126038 URL |
| [6] |
Suárez-López P, Wheatley K, Robson F, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis[J]. Nature, 2001,410(6832):1116-1120.
pmid: 11323677 |
| [7] |
Pin PA, Benlloch R, Bonnet D, et al. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet[J]. Science, 2010,330(6009):1397-1400.
doi: 10.1126/science.1197004 URL |
| [8] |
Trevaskis B, Hemming MN, Dennis ES, et al. The molecular basis of vernalization-induced flowering in cereals[J]. Trends Plant Sci, 2007,12(8):352-357.
pmid: 17629542 |
| [9] |
Trevaskis B, Bagnall DJ, Ellis MH, et al. MADS box genes control vernalization-induced flowering in cereals[J]. Proc Natl Acad Sci USA, 2003,100(22):13099-13104.
doi: 10.1073/pnas.1635053100 URL |
| [10] |
Yan L, Loukoianov A, Blechl A, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization[J]. Science, 2004,303(5664):1640-1644.
doi: 10.1126/science.1094305 URL |
| [11] |
Abe M, Kobayashi Y, Yamamoto S, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex[J]. Science, 2005,309(5737):1052-1056.
doi: 10.1126/science.1115983 URL |
| [12] |
Schwartz C, Balasubramanian S, Warthmann N, et al. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana[J]. Genetics, 2009,183(2):723-732.
doi: 10.1534/genetics.109.104984 URL |
| [13] |
Kojima S, Takahashi Y, Kobayashi Y, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions[J]. Plant Cell Physiol, 2002,43(10):1096-1105.
pmid: 12407188 |
| [14] |
Lee R, et al. FLOWERING LOCUS T genes control onion bulb formation and flowering[J]. Nat Commun, 2013,4:2884.
doi: 10.1038/ncomms3884 URL |
| [15] |
Guo D, Li C, et al. Molecular cloning and functional analysis of the FLOWERING LOCUS T(FT)homolog GhFT1 from Gossypium hirsutum[J]. J Integr Plant Biol, 2015,57(6):522-533.
doi: 10.1111/jipb.12316 URL |
| [16] |
Lee C, Kim SJ, Jin S, et al. Genetic interactions reveal the antagonistic roles of FT / TSF and TFL1 in the determination of inflorescence meristem identity in Arabidopsis[J]. The Plant Journal, 2019,99(3):452-464.
doi: 10.1111/tpj.v99.3 URL |
| [17] | Miho KS, Rie KI, Chiaki OT, et al. TFL1-Like proteins in rice antagonize rice FT-Like protein in inflorescence development by competition for complex formation with 14-3-3 and FD[J]. Plant & Cell Physiology, 2018,59(3):458-468 |
| [18] |
Wu L, Liu DF, Wu JJ. et al. Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon[J]. Plant Cell, 2013,25(11):4363-4377.
doi: 10.1105/tpc.113.118620 URL |
| [19] | 陈锡, 赵德刚, 陈莹, 等. 高羊茅FaFT2基因克隆及表达分析[J]. 植物生理学报, 2017,5(8):1523-1531. |
| Chen X, Zhao DG, et al. Cloning and analysis of FaFT2 gene in tall fescue[J]. Plant Physiol J, 2017,5(8):1523-1531. | |
| [20] |
Kong FJ, Liu BH, Xia ZJ, et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean[J]. Plant Physiology, 2010,154(3):1220-1231.
doi: 10.1104/pp.110.160796 URL |
| [21] |
Ken-ichiro T, Izuru O, Hiroyuki T, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen[J]. Nature, 2011,476(7360):332-335
doi: 10.1038/nature10272 pmid: 21804566 |
| [22] | 蔡宇鹏. 大豆FT同源基因GmFT2a和GmFT5a在花期调控中的遗传效应研究[D]. 北京:中国农业科学院, 2019. |
| Cai YP. Genetic effects of GmFT2a and GmFT5a on floral induction in soybean[D]. Beijing:Chinese Academy of Agricultural Sciences, 2019. |
| [1] | ZHANG Bin, YANG Xin-xia. Identification of Key Transcription Factors in Response to Salt Stress in Rice [J]. Biotechnology Bulletin, 2022, 38(3): 9-15. |
| [2] | DONG Hai-jiao, YANG Xiao-yu, MO Bei-xin, CHEN Xue-mei, CUI Jie. Research Progress in NAD+ Cap Modification at the 5' End of RNA [J]. Biotechnology Bulletin, 2022, 38(2): 245-251. |
| [3] | ZHAO Jie, LI An, LIANG Gang, JIN Xin-xin, PAN Li-gang. Research Progress in the Biological Functions of Plant circRNAs [J]. Biotechnology Bulletin, 2022, 38(10): 1-9. |
| [4] | WANG Zhi-shan, LI Ni, WANG Wei-ping, LIU Yang. Research Progress in Endophytic Bacteria in Rice Seeds [J]. Biotechnology Bulletin, 2022, 38(1): 236-246. |
| [5] | HE Xiao-li, GUO Lei-zhou, HAN Jia-hui, TANG Yin, YUAN Yuan, DAI Qi-lin, PING Shu-zhen, JIANG Shi-jie. Research Progress on Bacterial Periplasmic Chaperone LolA [J]. Biotechnology Bulletin, 2021, 37(8): 275-283. |
| [6] | XUE Xiang-lan, DING Yang-yang, LIU Yue, LI Xiao-bo, JIANG Lin, HE Xiao-hong, MA Yue-hui, ZHAO Qian-jun. Research Progress on Biological Function Growth and Development Related to N6-methyladenosine in Mammals [J]. Biotechnology Bulletin, 2021, 37(4): 251-259. |
| [7] | CHEN Ying, CHEN Xi, WANG Qian, WANG Xiao-li. Cloning,Expression and Biological Function Analysis of Universal Stress Protein in Festuca arundinacea [J]. Biotechnology Bulletin, 2021, 37(2): 32-39. |
| [8] | FENG Yi-long, ZHANG Wen-li. Research Progress on DNA Guanine Quadruplex [J]. Biotechnology Bulletin, 2020, 36(7): 23-31. |
| [9] | XU Lin-na, HU Meng-ke, TONG Wen-yan, LI Fen. Effects of T1084d and T1084A Point Mutations in the NtTkr Tail of Nicotiana tabacum on Coiled-helix Structure and Interaction with Target Proteins [J]. Biotechnology Bulletin, 2019, 35(5): 64-69. |
| [10] | JIA Jian-lei, CHEN Qian, JIN Ji-peng, YUAN Zan, ZHANG Li-ping. Eukaryotic Expression of BMPR1B in Sheep and Identification of Its Interaction Proteins [J]. Biotechnology Bulletin, 2019, 35(12): 94-104. |
| [11] | HUANG Xing, DING Feng, PENG Hong-xiang, PAN Jie-chun, HE Xin-hua, XU Jiong-zhi, LI Lin. Research Progress on Family of Plant WRKY Transcription Factors [J]. Biotechnology Bulletin, 2019, 35(12): 129-143. |
| [12] | WANG Chun-yu, ZHANG Qian. Research Progress on Plant NAC Transcription Factors [J]. Biotechnology Bulletin, 2018, 34(11): 8-14. |
| [13] | XU Hai-dong, LENG Qi-ying, PATRICIA Adu-Asiamah, WANG Zhang, LI Ting, ZHANG Li. Circular RNAs:Research Progress and Its Significance in Birds and Livestock [J]. Biotechnology Bulletin, 2018, 34(11): 56-69. |
| [14] | QIU Yan-hong,WANG Chao-nan,ZHU Shui-fang. Research Advances on the Pathogenicity of Cucumber Mosaic Virus [J]. Biotechnology Bulletin, 2017, 33(9): 10-16. |
| [15] | ZHANG Ying-yue ,MA Yue-hui ,ZHAO Qian-jun. Study Progress on Circular RNA [J]. Biotechnology Bulletin, 2017, 33(7): 29-34. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||