








生物技术通报 ›› 2025, Vol. 41 ›› Issue (5): 1-13.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1218
• 综述与专论 • 下一篇
收稿日期:2024-12-17
出版日期:2025-05-26
发布日期:2025-06-05
通讯作者:
高振宇,男,博士,研究员,研究方向 :作物遗传育种;E-mail: gaozhenyu@caas.cn作者简介:刘园园,女,硕士研究生,研究方向 :遗传学;E-mail: 1538389544@163.com
基金资助:
LIU Yuan-yuan1( ), CHEN Xi-feng1, QIAN Qian2,3, GAO Zhen-yu2(
), CHEN Xi-feng1, QIAN Qian2,3, GAO Zhen-yu2( )
)
Received:2024-12-17
Published:2025-05-26
Online:2025-06-05
摘要:
水稻是全球主要的粮食作物之一,为全球超过半数人口的基本食粮。有效穗数、穗粒数和千粒重是水稻产量的三大要素,水稻穗发育又是决定穗粒数的重要过程,并与产量的形成有着密切关系。因此,研究水稻穗发育及其分子调控机制可为水稻高产育种实践提供理论依据和指导。水稻穗发育的遗传机制和分子调控一直以来就受到育种家们的关注。本文基于国内外对水稻穗发育相关基因及其作用机理的研究进展以及近年来新发现的水稻穗发育调控基因,从水稻分生组织活性维持、花序分生组织向小穗分生组织转换、花器官形成过程和遗传调控方面展开论述。分类介绍了这些基因调控穗发育的分子机制及可能存在的相互作用关系,同时归纳了细胞分裂素、生长素、赤霉素、油菜素内酯等植物激素的信号途径中这些基因如何参与调控水稻穗发育,还进一步探讨了温度、光照、水分和营养元素等环境因素在水稻穗发育过程中的重要作用。在对水稻穗发育的分子调控机制和遗传调控网络展开全面综述的基础上,系统梳理和分析了现阶段水稻穗发育研究存在的棘手问题和相应的解决策略,并就未来水稻穗发育研究领域的关键问题和核心手段作了展望。
刘园园, 陈析丰, 钱前, 高振宇. 水稻穗发育调控的分子机制研究进展[J]. 生物技术通报, 2025, 41(5): 1-13.
LIU Yuan-yuan, CHEN Xi-feng, QIAN Qian, GAO Zhen-yu. Advances in Molecular Mechanisms Regulating Panicle Development in Rice[J]. Biotechnology Bulletin, 2025, 41(5): 1-13.

图1 水稻穗型发育模式图LP:叶原基;AM:腋生分生组织;SAM:茎顶端分生组织;RM:穗轴分生组织;MA:穗轴;PBM:一级枝梗分生组织;PB:一级枝梗;SBM:二级枝梗分生组织;SB:二级枝梗;TB:三级枝梗;TSM:终端小穗分生组织;TS:终端小穗;LSM:侧生小穗分生组织;LS:侧生小穗;RG:退化颖壳;P:内稃;L:外稃;EG:护颖;DP:退化点;:正调节;:负调节
Fig. 1 Rice panicle development modelLP: Leaf primordium; AM: axillary meristem; SAM: shoot apical meristem; RM: rachis meristem; MA: main axis; PBM: primary branch meristem; PB: primary branch; SBM: secondary branch meristem; SB: secondary branch; TB: tertiary branch; TSM: terminal spikelet meristem; TS: terminal spikelet; LSM: lateral spikelet meristem; LS: lateral spikelet; RG: retrogressive glume; P: palea; L: lemma; EG: empty glume; DP: degenerate point; : positive regulation; : negative regulation
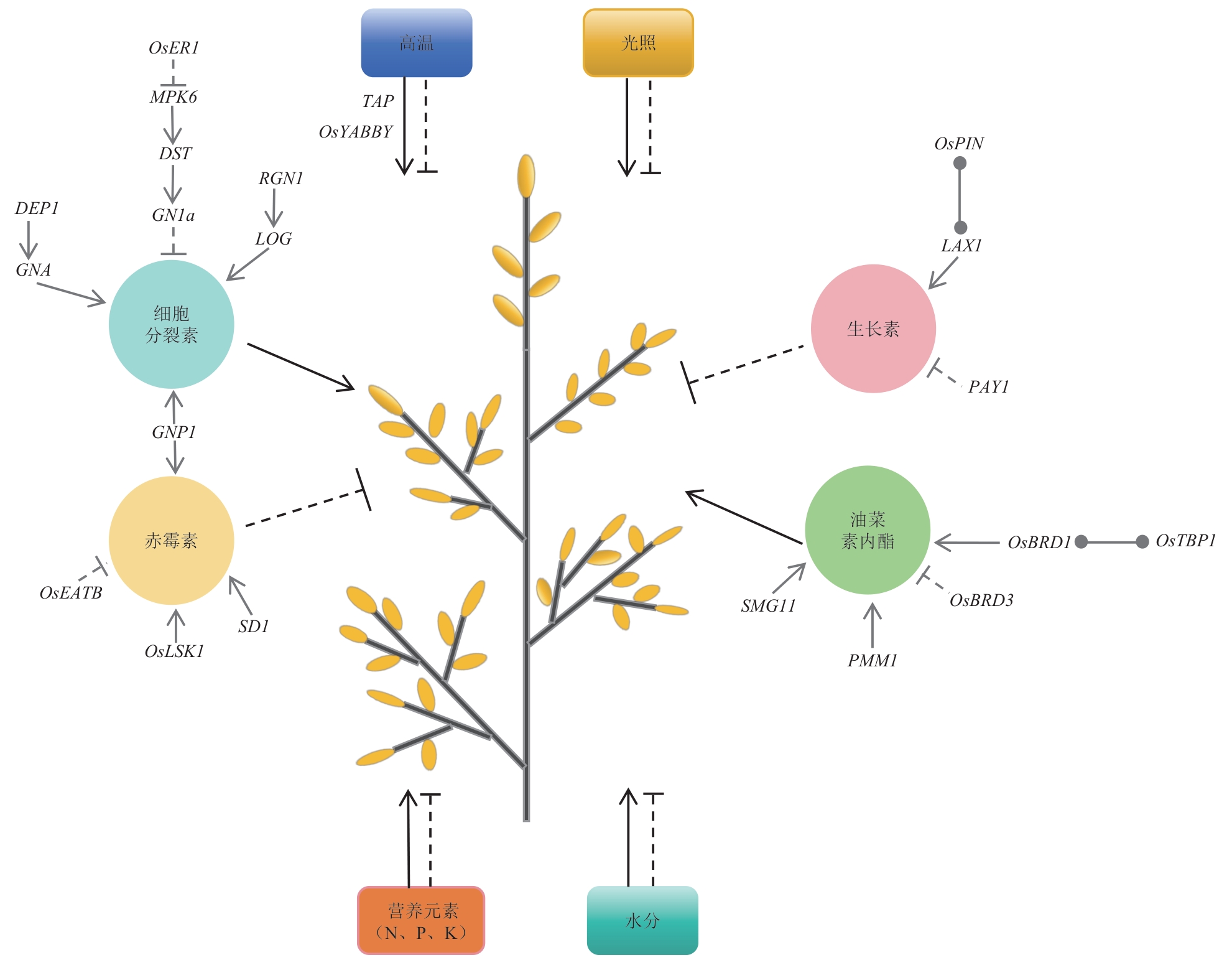
图2 激素和环境因素对水稻穗发育的影响:正调节;:负调节;:相互作用
Fig. 2 Effects of phytohormone and environmental factors on panicle development of rice: Positive regulation; : negative regulation; : interaction
基因/QTL Gene/QTL | 基因登录号 Loc number | 蛋白类型 Protein type | 功能 Function | 参考文献 Reference |
|---|---|---|---|---|
| FON1 | LOC_Os06g50340 | 富亮氨酸重复受体激酶 | 调节分生组织大小来控制营养和生殖发育 | [ |
| FON4 | LOC_Os11g38270 | 含CLE功能域的分泌蛋白 | 调节水稻茎尖分生组织大小和花分生组织的确定性 | [ |
| FCP1 | LOC_Os04g39770 | 含CLE结构域的分泌蛋白 | 调节茎尖分生组织和根顶端分生组织分生活性的维持 | [ |
| FOS1 | LOC_Os02g21890 | 含CLE结构域的分泌蛋白 | 控制水稻花器官数目 | [ |
| APO1 | LOC_Os06g45460 | F-box蛋白 | 参与分生组织的调控,正调节一级枝梗数目和小穗数目 | [ |
| APO2 | LOC_Os04g51000 | 拟南芥RFL同源蛋白 | 穗分枝形成的正调控因子,参与枝梗分生组织命运维持 | [ |
| TAW1 | LOC_Os10g33780 | ALOG家族蛋白 | 编码功能未知核蛋白,参与调控枝梗分生组织 | [ |
| DEP1 | LOC_Os09g26999 | 异三聚体G蛋白γ亚基 | 参与细胞分裂调控枝梗数和穗粒数 | [ |
| LAX1 | LOC_Os01g61480 | bHLH转录因子 | 控制水稻腋芽原基形成和小穗枝梗原基的分化 | [ |
| LAX2 | LOC_Os04g32510 | 核定位蛋白 | 含植物特异保守结构域,参与调控水稻腋生分生组织的形成 | [ |
| DST | LOC_Os03g57240 | 锌指转录因子 | 参与水稻穗部形态建成,负调控穗粒数 | [ |
| FZP | LOC_Os07g47330 | ERF转录因子 | 决定穗分枝向小穗形成的转化 | [ |
| SNB | LOC_Os07g13170 | 植物特有的AP2转录因子 | 参与小穗分生组织向花分生组织转变及花器官模式形成 | [ |
| OsIDS1 | LOC_Os03g60430 | AP2/ERF型转录抑制因子 | 调控水稻小穗发育和花器官特征 | [ |
| OsMFS1 | LOC_Os09g10850 | 减数分裂螺旋蛋白 | 主要在花药中表达,与小穗原基向花原基的转化有关 | [ |
| OsMADS34 | LOC_Os03g54170 | 属于SEP亚家族转录因子 | 调控水稻从营养生长向生殖生长转化及花序分生组织建成 | [ |
| OsMADS5 | LOC_Os06g06750 | 属于SEP亚家族转录因子 | 调控水稻穗发育,限制穗分枝及促进小穗分生组织特性转变 | [ |
| OsMADS1 | LOC_Os03g11614 | 编码由257个氨基酸组成的蛋白 | 参与花器官形成并抑制小穗分生组织的逆转 | [ |
| OsMADS16 | LOC_Os06g49840 | 含MADS框225个氨基酸组成的蛋白 | 控制水稻花器官的发育,调控浆片和雄蕊的发育 | [ |
| LOG | LOC_Os01g40630 | 细胞分裂素激活酶 | 与分生组织活性维持有关 | [ |
| An-1 | LOC_Os04g28280 | bHLH蛋白 | 调控水稻芒发育、籽粒大小和籽粒数 | [ |
| Gn1a | LOC_Os01g10110 | 降解细胞分裂素的酶 | 使细胞分裂素积累在花序分生组织中影响穗粒数 | [ |
| LP | LOC_Os02g15950 | 富含Kelch的F-box蛋白 | 参与植物组织的细胞分裂素水平的调节 | [ |
| SP3 | LOC_Os03g55610 | Dof转录因子 | 通过增加细胞分裂素含量控制水稻穗型 | [ |
| OsER1 | LOC_Os06g10230 | 类受体蛋白激酶 | 负调控穗粒数 | [ |
| OsPIN5b | LOC_Os08g41720 | PIN蛋白 | 改变生长素稳态、运输和分布,调控水稻株型和产量 | [ |
| OsPID | LOC_Os12g42020 | 含有激酶结构域蛋白 | 负调控穗粒数 | [ |
| PAY1 | LOC_Os08g31470 | 包含肽酶S64结构域蛋白 | 通过影响生长素极性运输改变穗粒数和产量 | [ |
| OsGRF6 | LOC_Os03g51970 | 生长调节因子 | 正调控生长素信号通路,促进花序发育,增加穗粒数 | [ |
| OsLSK1 | LOC_Os01g47900 | 典型的Ⅲ型S-结构域类受体激酶 | 影响株高和一次枝梗数和枝梗着粒数 | [ |
| EUI1 | LOC_Os05g40384 | 577个氨基酸组成的蛋白 | 促进细胞生长和细胞增殖来调节籽粒大小和剑叶夹角 | [ |
| OsBRD1 | LOC_Os03g40540 | BR生物合成关键酶 | 通过BR合成途径调控水稻穗和籽粒发育 | [ |
| SMG11 | LOC_Os01g10040 | 参与BR的生物合成的细胞色素 | 控制水稻籽粒大小、籽粒数目和谷物产量 | [ |
| OsTBP1 | LOC_Os08g07760 | BR信号受体BRI1的激酶 | 调节株高、叶夹角、穗粒数和籽粒大小 | [ |
| OsBRD3 | LOC_Os06g39880 | 编码BR代谢酶 | 参与调控二次枝梗分生组织的转变和二次枝梗的数目 | [ |
| TAP | LOC_Os02g18370 | 水稻转座酶衍生的转录因子蛋白 | 高温条件下能够介导调控水稻的花序维持和小穗正常发育 | [ |
| Ghd7 | LOC_Os07g15770 | 含CCT结构域的转录抑制因子 | 能同时控制水稻穗粒数、株高和抽穗期3个性状的主效QTL | [ |
表1 部分已克隆穗发育基因或QTL
Table 1 Some cloned genes or QTLs associated with panicle development
基因/QTL Gene/QTL | 基因登录号 Loc number | 蛋白类型 Protein type | 功能 Function | 参考文献 Reference |
|---|---|---|---|---|
| FON1 | LOC_Os06g50340 | 富亮氨酸重复受体激酶 | 调节分生组织大小来控制营养和生殖发育 | [ |
| FON4 | LOC_Os11g38270 | 含CLE功能域的分泌蛋白 | 调节水稻茎尖分生组织大小和花分生组织的确定性 | [ |
| FCP1 | LOC_Os04g39770 | 含CLE结构域的分泌蛋白 | 调节茎尖分生组织和根顶端分生组织分生活性的维持 | [ |
| FOS1 | LOC_Os02g21890 | 含CLE结构域的分泌蛋白 | 控制水稻花器官数目 | [ |
| APO1 | LOC_Os06g45460 | F-box蛋白 | 参与分生组织的调控,正调节一级枝梗数目和小穗数目 | [ |
| APO2 | LOC_Os04g51000 | 拟南芥RFL同源蛋白 | 穗分枝形成的正调控因子,参与枝梗分生组织命运维持 | [ |
| TAW1 | LOC_Os10g33780 | ALOG家族蛋白 | 编码功能未知核蛋白,参与调控枝梗分生组织 | [ |
| DEP1 | LOC_Os09g26999 | 异三聚体G蛋白γ亚基 | 参与细胞分裂调控枝梗数和穗粒数 | [ |
| LAX1 | LOC_Os01g61480 | bHLH转录因子 | 控制水稻腋芽原基形成和小穗枝梗原基的分化 | [ |
| LAX2 | LOC_Os04g32510 | 核定位蛋白 | 含植物特异保守结构域,参与调控水稻腋生分生组织的形成 | [ |
| DST | LOC_Os03g57240 | 锌指转录因子 | 参与水稻穗部形态建成,负调控穗粒数 | [ |
| FZP | LOC_Os07g47330 | ERF转录因子 | 决定穗分枝向小穗形成的转化 | [ |
| SNB | LOC_Os07g13170 | 植物特有的AP2转录因子 | 参与小穗分生组织向花分生组织转变及花器官模式形成 | [ |
| OsIDS1 | LOC_Os03g60430 | AP2/ERF型转录抑制因子 | 调控水稻小穗发育和花器官特征 | [ |
| OsMFS1 | LOC_Os09g10850 | 减数分裂螺旋蛋白 | 主要在花药中表达,与小穗原基向花原基的转化有关 | [ |
| OsMADS34 | LOC_Os03g54170 | 属于SEP亚家族转录因子 | 调控水稻从营养生长向生殖生长转化及花序分生组织建成 | [ |
| OsMADS5 | LOC_Os06g06750 | 属于SEP亚家族转录因子 | 调控水稻穗发育,限制穗分枝及促进小穗分生组织特性转变 | [ |
| OsMADS1 | LOC_Os03g11614 | 编码由257个氨基酸组成的蛋白 | 参与花器官形成并抑制小穗分生组织的逆转 | [ |
| OsMADS16 | LOC_Os06g49840 | 含MADS框225个氨基酸组成的蛋白 | 控制水稻花器官的发育,调控浆片和雄蕊的发育 | [ |
| LOG | LOC_Os01g40630 | 细胞分裂素激活酶 | 与分生组织活性维持有关 | [ |
| An-1 | LOC_Os04g28280 | bHLH蛋白 | 调控水稻芒发育、籽粒大小和籽粒数 | [ |
| Gn1a | LOC_Os01g10110 | 降解细胞分裂素的酶 | 使细胞分裂素积累在花序分生组织中影响穗粒数 | [ |
| LP | LOC_Os02g15950 | 富含Kelch的F-box蛋白 | 参与植物组织的细胞分裂素水平的调节 | [ |
| SP3 | LOC_Os03g55610 | Dof转录因子 | 通过增加细胞分裂素含量控制水稻穗型 | [ |
| OsER1 | LOC_Os06g10230 | 类受体蛋白激酶 | 负调控穗粒数 | [ |
| OsPIN5b | LOC_Os08g41720 | PIN蛋白 | 改变生长素稳态、运输和分布,调控水稻株型和产量 | [ |
| OsPID | LOC_Os12g42020 | 含有激酶结构域蛋白 | 负调控穗粒数 | [ |
| PAY1 | LOC_Os08g31470 | 包含肽酶S64结构域蛋白 | 通过影响生长素极性运输改变穗粒数和产量 | [ |
| OsGRF6 | LOC_Os03g51970 | 生长调节因子 | 正调控生长素信号通路,促进花序发育,增加穗粒数 | [ |
| OsLSK1 | LOC_Os01g47900 | 典型的Ⅲ型S-结构域类受体激酶 | 影响株高和一次枝梗数和枝梗着粒数 | [ |
| EUI1 | LOC_Os05g40384 | 577个氨基酸组成的蛋白 | 促进细胞生长和细胞增殖来调节籽粒大小和剑叶夹角 | [ |
| OsBRD1 | LOC_Os03g40540 | BR生物合成关键酶 | 通过BR合成途径调控水稻穗和籽粒发育 | [ |
| SMG11 | LOC_Os01g10040 | 参与BR的生物合成的细胞色素 | 控制水稻籽粒大小、籽粒数目和谷物产量 | [ |
| OsTBP1 | LOC_Os08g07760 | BR信号受体BRI1的激酶 | 调节株高、叶夹角、穗粒数和籽粒大小 | [ |
| OsBRD3 | LOC_Os06g39880 | 编码BR代谢酶 | 参与调控二次枝梗分生组织的转变和二次枝梗的数目 | [ |
| TAP | LOC_Os02g18370 | 水稻转座酶衍生的转录因子蛋白 | 高温条件下能够介导调控水稻的花序维持和小穗正常发育 | [ |
| Ghd7 | LOC_Os07g15770 | 含CCT结构域的转录抑制因子 | 能同时控制水稻穗粒数、株高和抽穗期3个性状的主效QTL | [ |
| 1 | Zuo JR, Li JY. Molecular genetic dissection of quantitative trait loci regulating rice grain size [J]. Annu Rev Genet, 2014, 48: 99-118. |
| 2 | Li GL, Zhang HL, Li JJ, et al. Genetic control of panicle architecture in rice [J]. Crop J, 2021, 9(3): 590-597. |
| 3 | 卢寰, 时振英. 水稻穗发育的分子生物学研究进展 [J]. 植物生理学报, 2013, 49(2): 111-121. |
| Lu H, Shi ZY. Molecular research progress of rice panicle development [J]. Plant Physiol J, 2013, 49(2): 111-121. | |
| 4 | Ikeda K, Sunohara H, Nagato Y. Developmental course of inflorescence and spikelet in rice [J]. Breed Sci, 2004, 54(2): 147-156. |
| 5 | Itoh JI, Nonomura KI, Ikeda K, et al. Rice plant development: from zygote to spikelet [J]. Plant Cell Physiol, 2005, 46(1): 23-47. |
| 6 | Moon S, Jung KH, Lee DE, et al. The rice FON1 gene controls vegetative and reproductive development by regulating shoot apical meristem size [J]. Mol Cells, 2006, 21(1): 147-152. |
| 7 | Suzaki T, Toriba T, Fujimoto M, et al. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene [J]. Plant Cell Physiol, 2006, 47(12): 1591-1602. |
| 8 | Chu HW, Qian Q, Liang WQ, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice [J]. Plant Physiol, 2006, 142(3): 1039-1052. |
| 9 | Ren DY, Xu QK, Qiu ZN, et al. FON4 prevents the multi-floret spikelet in rice [J]. Plant Biotechnol J, 2019, 17(6): 1007-1009. |
| 10 | Ohmori Y, Tanaka W, Kojima M, et al. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice [J]. Plant Cell, 2013, 25(1): 229-241. |
| 11 | Suzaki T, Ohneda M, Toriba T, et al. FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice [J]. PLoS Genet, 2009, 5(10): e1000693. |
| 12 | Ikeda-Kawakatsu K, Yasuno N, Oikawa T, et al. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem [J]. Plant Physiol, 2009, 150(2): 736-747. |
| 13 | Ikeda-Kawakatsu K, Maekawa M, Izawa T, et al. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1 [J]. Plant J, 2012, 69(1): 168-180. |
| 14 | Huang LJ, Hua K, Xu R, et al. The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice [J]. Plant Cell, 2021, 33(4): 1212-1228. |
| 15 | Yoshida A, Sasao M, Yasuno N, et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition [J]. Proc Natl Acad Sci USA, 2013, 110(2): 767-772. |
| 16 | Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice [J]. Plant J, 2002, 29(6): 743-750. |
| 17 | Huang XZ, Qian Q, Liu ZB, et al. Natural variation at the DEP1 locus enhances grain yield in rice [J]. Nat Genet, 2009, 41(4): 494-497. |
| 18 | Li F, Liu WB, Tang JY, et al. Rice dense and erect panicle 2 is essential for determining panicle outgrowth and elongation [J]. Cell Res, 2010, 20(7): 838-849. |
| 19 | Matin MN, Kang SG. Genetic and phenotypic analysis of lax1-6, a mutant allele of LAX PANICLE1 in rice [J]. J Plant Biol, 2012, 55(1): 50-63. |
| 20 | Tabuchi H, Zhang Y, Hattori S, et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems [J]. Plant Cell, 2011, 23(9): 3276-3287. |
| 21 | Wu HM, Xie DJ, Tang ZS, et al. PINOID regulates floral organ development by modulating auxin transport and interacts with MADS16 in rice [J]. Plant Biotechnol J, 2020, 18(8): 1778-1795. |
| 22 | Han ML, Lv QY, Zhang J, et al. Decreasing nitrogen assimilation under drought stress by suppressing DST-mediated activation of Nitrate Reductase 1.2 in rice [J]. Mol Plant, 2022, 15(1): 167-178. |
| 23 | Bai XF, Huang Y, Mao DH, et al. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice [J]. Sci Rep, 2016, 6: 19022. |
| 24 | Bai XF, Huang Y, Hu Y, et al. Duplication of an upstream silencer of FZP increases grain yield in rice [J]. Nat Plants, 2017, 3(11): 885-893. |
| 25 | Jiang LY, Ma X, Zhao SS, et al. The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size [J]. Plant Cell, 2019, 31(1): 17-36. |
| 26 | Lee DY, An G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice [J]. Plant J, 2012, 69(3): 445-461. |
| 27 | Lu JY, Wang CL, Wang HY, et al. OsMFS1/OsHOP2 complex participates in rice male and female development [J]. Front Plant Sci, 2020, 11: 518. |
| 28 | Ren DY, Yu HP, Rao YC, et al. 'Two-floret spikelet' as a novel resource has the potential to increase rice yield [J]. Plant Biotechnol J, 2018, 16(2): 351-353. |
| 29 | Kobayashi K, Maekawa M, Miyao A, et al. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice [J]. Plant Cell Physiol, 2010, 51(1): 47-57. |
| 30 | Zhu WW, Yang L, Wu D, et al. Rice SEPALLATA genes OsMADS5 and OsMADS34 cooperate to limit inflorescence branching by repressing the TERMINAL FLOWER1-like gene RCN4 [J]. New Phytol, 2022, 233(4): 1682-1700. |
| 31 | Chung YY, Kim SR, Finkel D, et al. Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene [J]. Plant Mol Biol, 1994, 26(2): 657-665. |
| 32 | Jeon JS, Jang S, Lee S, et al. Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development [J]. Plant Cell, 2000, 12(6): 871-884. |
| 33 | Agrawal GK, Abe K, Yamazaki M, et al. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene [J]. Plant Mol Biol, 2005, 59(1): 125-135. |
| 34 | Wang L, Zeng XQ, Zhuang H, et al. Ectopic expression of OsMADS1 caused dwarfism and spikelet alteration in rice [J]. Plant Growth Regul, 2017, 81(3): 433-442. |
| 35 | Xiao H, Wang Y, Liu DF, et al. Functional analysis of the rice AP3 homologue OsMADS16 by RNA interference [J]. Plant Mol Biol, 2003, 52(5): 957-966. |
| 36 | Dreni L, Jacchia S, Fornara F, et al. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice [J]. Plant J, 2007, 52(4): 690-699. |
| 37 | Cui RF, Han JK, Zhao SZ, et al. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa) [J]. Plant J, 2010, 61(5): 767-781. |
| 38 | Pelucchi N, Fornara F, Favalli C, et al. Comparative analysis of rice MADS-box genes expressed during flower development [J]. Sex Plant Reprod, 2002, 15(3): 113-122. |
| 39 | Wang KJ, Tang D, Hong LL, et al. DEP and AFO regulate reproductive habit in rice [J]. PLoS Genet, 2010, 6(1): e1000818. |
| 40 | Sang XC, Li YF, Luo ZK, et al. CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice[J]. Plant Physiol, 2012, 160(2): 788-807. |
| 41 | Hu Y, Wang L, Jia R, et al. Rice transcription factor MADS32 regulates floral patterning through interactions with multiple floral homeotic genes [J]. J Exp Bot, 2021, 72(7): 2434-2449. |
| 42 | Rashotte AM. The evolution of cytokinin signaling and its role in development before Angiosperms [J]. Semin Cell Dev Biol, 2021, 109: 31-38. |
| 43 | Kurakawa T, Ueda N, Maekawa M, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme [J]. Nature, 2007, 445(7128): 652-655. |
| 44 | Li GL, Xu BX, Zhang YP, et al. RGN1 controls grain number and shapes panicle architecture in rice [J]. Plant Biotechnol J, 2022, 20(1): 158-167. |
| 45 | Gu BG, Zhou TY, Luo JH, et al. An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice [J]. Mol Plant, 2015, 8(11): 1635-1650. |
| 46 | Ashikari M, Sakakibara H, Lin SY, et al. Cytokinin oxidase regulates rice grain production [J]. Science, 2005, 309(5735): 741-745. |
| 47 | Li SY, Zhao BR, Yuan DY, et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression [J]. Proc Natl Acad Sci USA, 2013, 110(8): 3167-3172. |
| 48 | Li M, Tang D, Wang KJ, et al. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice [J]. Plant Biotechnol J, 2011, 9(9): 1002-1013. |
| 49 | Huang Y, Bai XF, Luo MF, et al. Short Panicle 3 controls panicle architecture by upregulating APO2/RFL and increasing cytokinin content in rice [J]. J Integr Plant Biol, 2019, 61(9): 987-999. |
| 50 | Wu Y, Wang Y, Mi XF, et al. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems [J]. PLoS Genet, 2016, 12(10): e1006386. |
| 51 | Rong CY, Liu YX, Chang ZY, et al. Cytokinin oxidase/dehydrogenase family genes exhibit functional divergence and overlap in rice growth and development, especially in control of tillering [J]. J Exp Bot, 2022, 73(11): 3552-3568. |
| 52 | Guo T, Lu ZQ, Shan JX, et al. ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice [J]. Plant Cell, 2020, 32(9): 2763-2779. |
| 53 | Zhang JH, Lin QB, Wang X, et al. The dense and erect panicle1-grain number associated module enhances rice yield by repressing cytokinin oxidase 2 expression [J]. Plant Cell, 2024, 37(1): koae309. |
| 54 | 淳雁, 李学勇. 水稻穗型的遗传调控研究进展 [J]. 植物学报, 2017, 52(1): 19-29. |
| Chun Y, Li XY. Research progress in genetic regulation of rice panicle architecture [J]. Chin Bull Bot, 2017, 52(1): 19-29. | |
| 55 | Lu GW, Coneva V, Casaretto JA, et al. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution [J]. Plant J, 2015, 83(5): 913-925. |
| 56 | Morita Y, Kyozuka J. Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport [J]. Plant Cell Physiol, 2007, 48(3): 540-549. |
| 57 | Zhao L, Tan LB, Zhu ZF, et al. PAY1 improves plant architecture and enhances grain yield in rice [J]. Plant J, 2015, 83(3): 528-536. |
| 58 | Tang YY, Liu HH, Guo SY, et al. OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice [J]. Plant Physiol, 2018, 176(1): 946-959. |
| 59 | Gao SP, Chu CC. Gibberellin metabolism and signaling: targets for improving agronomic performance of crops [J]. Plant Cell Physiol, 2020, 61(11): 1902-1911. |
| 60 | Agata A, Ando K, Ota S, et al. Diverse panicle architecture results from various combinations of Prl5/GA20ox4 and Pbl6/APO1 alleles [J]. Commun Biol, 2020, 3(1): 302. |
| 61 | Su S, Hong J, Chen XF, et al. Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice [J]. Plant Biotechnol J, 2021, 19(11): 2304-2318. |
| 62 | Qi WW, Sun F, Wang QJ, et al. Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene [J]. Plant Physiol, 2011, 157(1): 216-228. |
| 63 | Zou XH, Qin ZR, Zhang CY, et al. Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice [J]. J Exp Bot, 2015, 66(22): 7197-7209. |
| 64 | 肖辉海. 水稻长穗颈隐性高秆突变体穗颈节间的细胞学观察 [J]. 西北农林科技大学学报: 自然科学版, 2008, 36(1): 131-136. |
| Xiao HH. Cytological studies on uppermost internode of recessive tall stalk rice with eui gene [J]. J Northwest A F Univ Nat Sci Ed, 2008, 36(1): 131-136. | |
| 65 | Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem [J]. Plant J, 2002, 32(4): 495-508. |
| 66 | Hong Z, Ueguchi-Tanaka M, Fujioka S, et al. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone [J]. Plant Cell, 2005, 17(8): 2243-2254. |
| 67 | Li Y, Li XM, Fu DB, et al. Panicle Morphology Mutant 1 (PMM1) determines the inflorescence architecture of rice by controlling brassinosteroid biosynthesis [J]. BMC Plant Biol, 2018, 18(1): 348. |
| 68 | Fang N, Xu R, Huang LJ, et al. SMALL GRAIN 11 controls grain size, grain number and grain yield in rice [J]. Rice, 2016, 9(1): 64. |
| 69 | Chen XW, Zuo SM, Schwessinger B, et al. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors [J]. Mol Plant, 2014, 7(5): 874-892. |
| 70 | Lin Y, Zhao ZG, Zhou SR, et al. Top Bending Panicle1 is involved in brassinosteroid signaling and regulates the plant architecture in rice [J]. Plant Physiol Biochem, 2017, 121: 1-13. |
| 71 | Zhang XX, Meng WJ, Liu DP, et al. Enhancing rice panicle branching and grain yield through tissue-specific brassinosteroid inhibition [J]. Science, 2024, 383(6687): eadk8838. |
| 72 | Zhang P, Zhu WW, He Y, et al. THERMOSENSITIVE BARREN PANICLE (TAP) is required for rice panicle and spikelet development at high ambient temperature [J]. New Phytol, 2023, 237(3): 855-869. |
| 73 | 成勤勤. 关于水稻幼穗分化发育的研究 [D]. 扬州: 扬州大学, 2006. |
| Cheng QQ. Study on the differentiation and development of young panicles in rice [D]. Yangzhou: Yangzhou University, 2006. | |
| 74 | Zhang WY, Chen YJ, Wang ZQ, et al. Polyamines and ethylene in rice young panicles in response to soil drought during panicle differentiation [J]. Plant Growth Regul, 2017, 82(3): 491-503. |
| 75 | Ding CQ, You J, Chen L, et al. Nitrogen fertilizer increases spikelet number per panicle by enhancing cytokinin synthesis in rice [J]. Plant Cell Rep, 2014, 33(2): 363-371. |
| 76 | Chen Q, Tian FA, Cheng TT, et al. Translational repression of FZP mediated by CU-rich element/OsPTB interactions modulates panicle development in rice [J]. Plant J, 2022, 110(5): 1319-1331. |
| 77 | Wu LH, Hu M, Lyu SW, et al. A 48-bp deletion upstream of LIGULELESS 1 alters rice panicle architecture [J]. Crop J, 2024, 12(2): 354-363. |
| 78 | Wu XW, Liang Y, Gao HB, et al. Enhancing rice grain production by manipulating the naturally evolved cis-regulatory element-containing inverted repeat sequence of OsREM20 [J]. Mol Plant, 2021, 14(6): 997-1011. |
| 79 | Xue WY, Xing YZ, Weng XY, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice [J]. Nat Genet, 2008, 40(6): 761-767. |
| 80 | Si LZ, Chen JY, Huang XH, et al. OsSPL13 controls grain size in cultivated rice [J]. Nat Genet, 2016, 48(4): 447-456. |
| 81 | Peng YL, Gao ZY, Zhang B, et al. Fine mapping and candidate gene analysis of a major QTL for panicle structure in rice [J]. Plant Cell Rep, 2014, 33(11): 1843-1850. |
| 82 | Liu EB, Liu Y, Wu GC, et al. Identification of a candidate gene for panicle length in rice (Oryza sativa L.) via association and linkage analysis [J]. Front Plant Sci, 2016, 7: 596. |
| 83 | Yang Y, Zhang Y, Li J, et al. Three QTL from Oryza meridionalis could improve panicle architecture in Asian cultivated rice [J]. Rice, 2023, 16(1): 22. |
| [1] | 胡若群, 曾菁菁, 梁婉凤, 曹佳玉, 黄小苇, 梁晓英, 仇明月, 陈莹. 转录组和代谢组联合分析探究不同遮光条件下金线莲类胡萝卜素合成代谢机制[J]. 生物技术通报, 2025, 41(5): 231-243. |
| [2] | 杜量衡, 唐黄磊, 张治国. 控制水稻光响应基因ELM1的图位克隆[J]. 生物技术通报, 2025, 41(5): 82-89. |
| [3] | 陈晓军, 惠建, 马洪文, 白海波, 钟楠, 李稼润, 樊云芳. 利用单碱基基因编辑技术创制OsALS抗除草剂水稻种质资源[J]. 生物技术通报, 2025, 41(4): 106-114. |
| [4] | 李欣芃, 张武汉, 张莉, 舒服, 何强, 郭杨, 邓华凤, 王悦, 孙平勇. γ射线诱变创制水稻突变体及其分子鉴定[J]. 生物技术通报, 2025, 41(3): 35-43. |
| [5] | 李艳伟, 杨妍妍, 孙亚玲, 霍雨猛, 王振宝, 刘冰江. 基于转录组分析植物激素对洋葱鳞茎膨大发育的调控机制[J]. 生物技术通报, 2025, 41(2): 187-201. |
| [6] | 匡健华, 程志鹏, 赵永晶, 杨洁, 陈润乔, 陈龙清, 胡慧贞. 激素和非生物胁迫下荷花GH3基因家族的表达分析[J]. 生物技术通报, 2025, 41(2): 221-233. |
| [7] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [8] | 金素奎, 国倩倩, 刘巧泉, 高继平. 一种水稻叶片基因组DNA简易提取方法[J]. 生物技术通报, 2025, 41(1): 74-84. |
| [9] | 刘文志, 贺丹, 李鹏, 傅应林, 张译心, 温华杰, 于文清. 多粘类芽胞杆菌新菌株X-11及其对番茄和水稻的促生效应[J]. 生物技术通报, 2024, 40(9): 249-259. |
| [10] | 朱诗斐, 刘敬, 张家芊, 黄文坤, 彭德良, 孔令安, 彭焕. 水稻和拟禾本科根结线虫互作分子机制研究进展[J]. 生物技术通报, 2024, 40(9): 172-180. |
| [11] | 李庆懋, 彭聪归, 齐笑含, 刘兴蕾, 李臻园, 李沁妍, 黄立钰. 促进水稻铁素吸收的野生稻内生细菌优良菌株的筛选与鉴定[J]. 生物技术通报, 2024, 40(8): 255-263. |
| [12] | 孙志勇, 杜怀东, 刘阳, 马嘉欣, 于雪然, 马伟, 姚鑫杰, 王敏, 李培富. 水稻籽粒γ-氨基丁酸含量的全基因组关联分析[J]. 生物技术通报, 2024, 40(8): 53-62. |
| [13] | 聂祝欣, 郭瑾, 乔子洋, 李微薇, 张学燕, 刘春阳, 王静. 黑果枸杞不同发育时期果实花色苷合成的转录组分析[J]. 生物技术通报, 2024, 40(8): 106-117. |
| [14] | 杜仲阳, 杨泽, 梁梦静, 刘义珍, 崔红利, 史达明, 薛金爱, 孙岩, 张春辉, 季春丽, 李润植. 纳米硒(SeNPs)缓解烟草幼苗铅胁迫和促生效应[J]. 生物技术通报, 2024, 40(7): 183-196. |
| [15] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||