








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 230-239.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1004
• 研究报告 • 上一篇
彭婷1,3( ), 林颖2, 谭圆圆1, 饶英1, 黄覃3, 张文娥1, 汪波4, 田瑞丰5, 刘国锋2(
), 林颖2, 谭圆圆1, 饶英1, 黄覃3, 张文娥1, 汪波4, 田瑞丰5, 刘国锋2( )
)
收稿日期:2024-10-15
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
刘国锋,男,博士,研究方向 :园林植物遗传育种;E-mail: gfliu@mail.hzau.edu.cn作者简介:彭婷,女,博士,研究方向 :园林植物遗传育种;E-mail: tpeng@mail.hzau.edu.cn
基金资助:
PENG Ting1,3( ), LIN Ying2, TAN Yuan-yuan1, RAO Ying1, HUANG Qin3, ZHANG Wen-e1, WANG Bo4, TIAN Rui-feng5, LIU Guo-feng2(
), LIN Ying2, TAN Yuan-yuan1, RAO Ying1, HUANG Qin3, ZHANG Wen-e1, WANG Bo4, TIAN Rui-feng5, LIU Guo-feng2( )
)
Received:2024-10-15
Published:2025-03-26
Online:2025-03-20
摘要:
目的 多星韭(Allium wallichii)是极具特色的高山花卉,花青苷合成酶(ANS)是类黄酮代谢途径下游合成花青苷的关键酶,研究ANS基因在紫花和白花多星韭花色形成中发挥的作用。 方法 采用pH示差法检测多星韭紫花和白花不同发育时期总花青苷含量;利用RT-PCR技术克隆紫花和白花AwANS基因,并对其进行表达模式分析。 结果 紫花多星韭花朵总花青苷含量随花发育逐渐增加,在S4达到峰值,白花花瓣中均检测不到花青苷含量。从紫花中克隆到2条ANS基因序列(AwANSpa 和AwANSpb ),CDS长度均为1 074 bp,编码357个氨基酸;从白花中克隆到5条ANS序列(AwANSwa 、AwANSwb 、AwANSwc 、AwANSwd 、AwANSwe ),AwANSwa 和AwANSwd CDS长度分别为1 074 bp和1 077 bp,分别编码357、358个氨基酸,另外3条序列出现大片段缺失导致蛋白翻译提前终止。系统进化树分析显示AwANSs与洋葱亲缘关系最近。RT-qPCR分析显示,紫花AwANSs在花瓣中表达量最高,在根、雌蕊和果实中表达量极低;AwANSs在紫花多星韭的表达随花发育而逐渐升高,在S5达到峰值;而在白花中几乎检测不到AwANSs表达。 结论 AwANSs的表达具有明显的时空表达特异性,在花瓣中表达量最高。与紫花相比,白花多星韭的花瓣中不积累花青素,其AwANSs基因在整个花发育过程中也几乎不表达,表明AwANSs基因对多星韭紫色花的形成起着重要作用。
彭婷, 林颖, 谭圆圆, 饶英, 黄覃, 张文娥, 汪波, 田瑞丰, 刘国锋. 多星韭AwANSs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(3): 230-239.
PENG Ting, LIN Ying, TAN Yuan-yuan, RAO Ying, HUANG Qin, ZHANG Wen-e, WANG Bo, TIAN Rui-feng, LIU Guo-feng. Cloning and Expression Analysis of AwANS Genes in Allium wallichii[J]. Biotechnology Bulletin, 2025, 41(3): 230-239.
引物名称 Primer name | 序列 Primer sequence(5′-3′) | 用途 Usage |
|---|---|---|
| AwANSs-Fw | AACTACAACATTACCTACAAACACAA | cDNA克隆 |
| AwANSs-Rev | TGTAGACGAACCAGGCTCAA | cDNA克隆 |
| AwANSs-qFw | TCACACCACCACCTGCAAGAGT | RT-qPCR |
| AwANSs-qRev | CGGATGAGCGAAGGTCGGTTATTG | RT-qPCR |
| AwTUB2-qFw | GAAAATCCGAGAAGAATACCCAGAC | RT-qPCR |
| AwTUB2-qRev | TGTCGTAAAGGGCTTCGTTGTC | RT-qPCR |
表1 本研究所用引物
Table 1 Primer sequences used in this study
引物名称 Primer name | 序列 Primer sequence(5′-3′) | 用途 Usage |
|---|---|---|
| AwANSs-Fw | AACTACAACATTACCTACAAACACAA | cDNA克隆 |
| AwANSs-Rev | TGTAGACGAACCAGGCTCAA | cDNA克隆 |
| AwANSs-qFw | TCACACCACCACCTGCAAGAGT | RT-qPCR |
| AwANSs-qRev | CGGATGAGCGAAGGTCGGTTATTG | RT-qPCR |
| AwTUB2-qFw | GAAAATCCGAGAAGAATACCCAGAC | RT-qPCR |
| AwTUB2-qRev | TGTCGTAAAGGGCTTCGTTGTC | RT-qPCR |
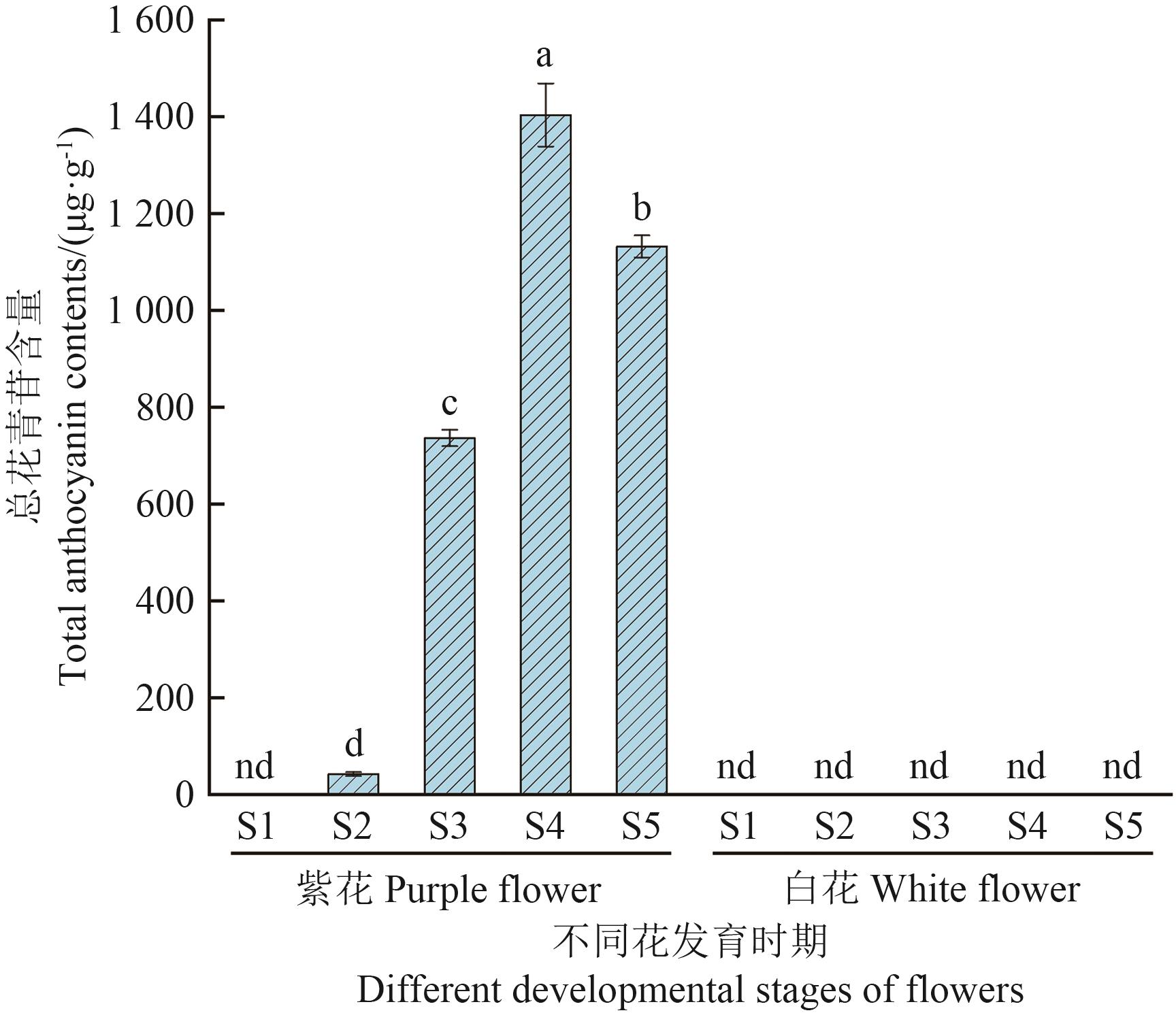
图2 多星韭不同花发育时期的总花青苷含量数据结果为3次生物学重复的平均值±标准误,采用Duncan多重检验,不同字母表示差异显著性(P<0.05),下同。“nd”表示“未检测到”
Fig. 2 Total anthocyanin contents at different developmental stages of flowers in A. wallichiiData are as mean±SE of three independent biological replicates. According to the Duncan multiple test, the different letters above each column indicate significant differences at 0.05 level. The same below. “nd” indicates that the total anthocyanin content is not detected
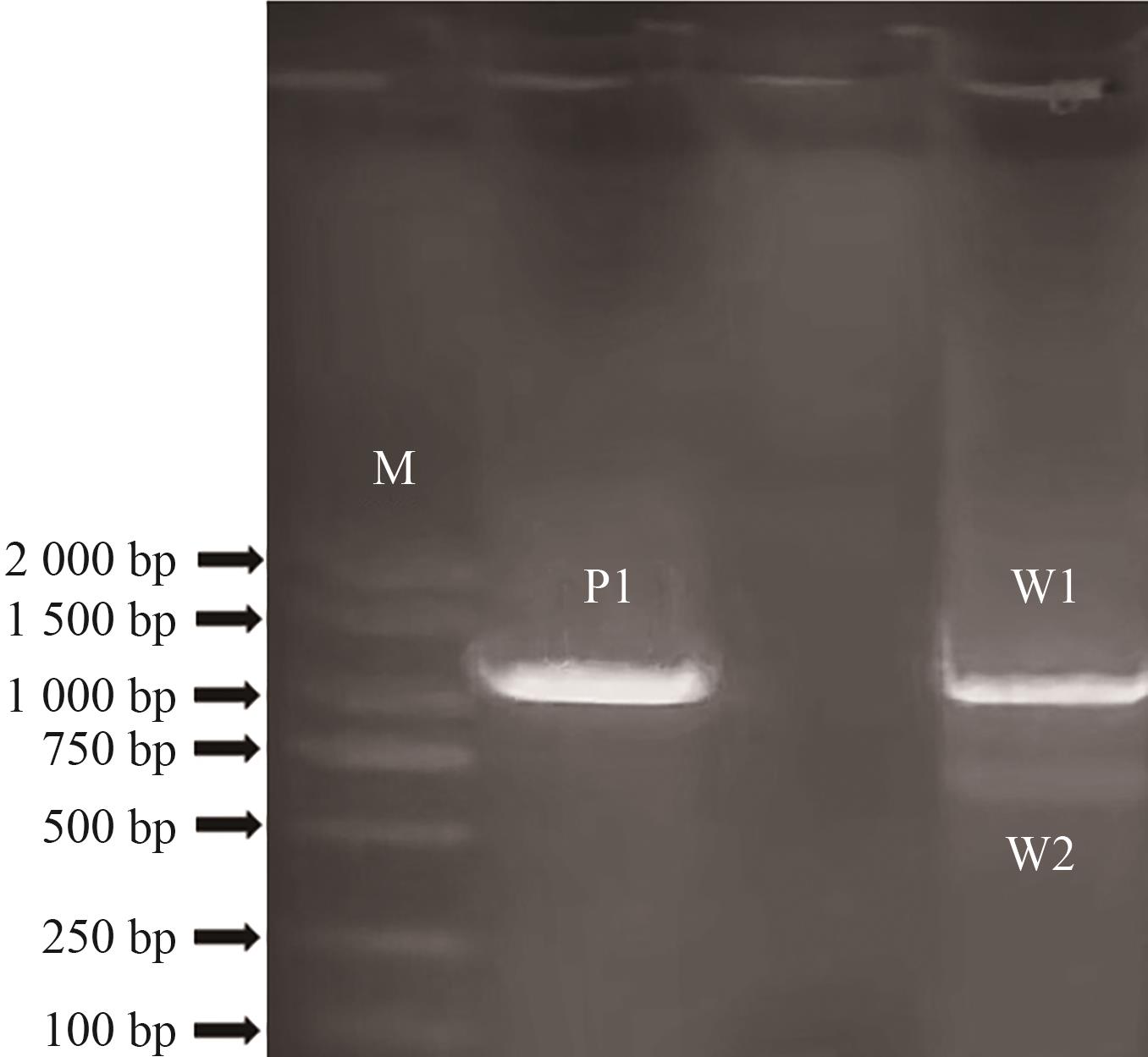
图3 AwANSs基因扩增M代表分子量标记,P1代表紫花多星韭,W1、W2代表白花多星韭
Fig. 3 Amplification of AwANSsM refers to marker. P1 indicates purple A. wallichii. W1 and W2 indicate white A. wallichii

图4 多星韭(A. wallichii)与其他物种ANS序列比对同源比对的各序列为水稻OsANS(CAA69252.1)、拟南芥AtANS(AT4G22880)、郁金香TgANS(USH59355.1)、洋葱AcANSL(ABM66367.1)。黑色线条标记为2-酮戊二酸双加氧酶家族保守结构域,*和#分别表示亚铁离子结合位点氨基酸和2-酮戊二酸结合位点氨基酸
Fig. 4 Sequence alignment of ANSs from A. wallichii and other speciesAwANSs were aligned with other ANSs from Oryza sativa (OsANS: CAA69252.1), Arabidopsis thaliana (AtANS: AT4G22880), Tulipa gesneriana (TgANS: USH59355.1), and Allium cepa (AcANSL: ABM66367.1). Conserved domain 2OG-Fe Ⅱ_Oxy is indicated by the solid line. Conserved residues that are required for ferrous-iron coordination and 2-oxoglutarate binding are marked by * and #, respectively
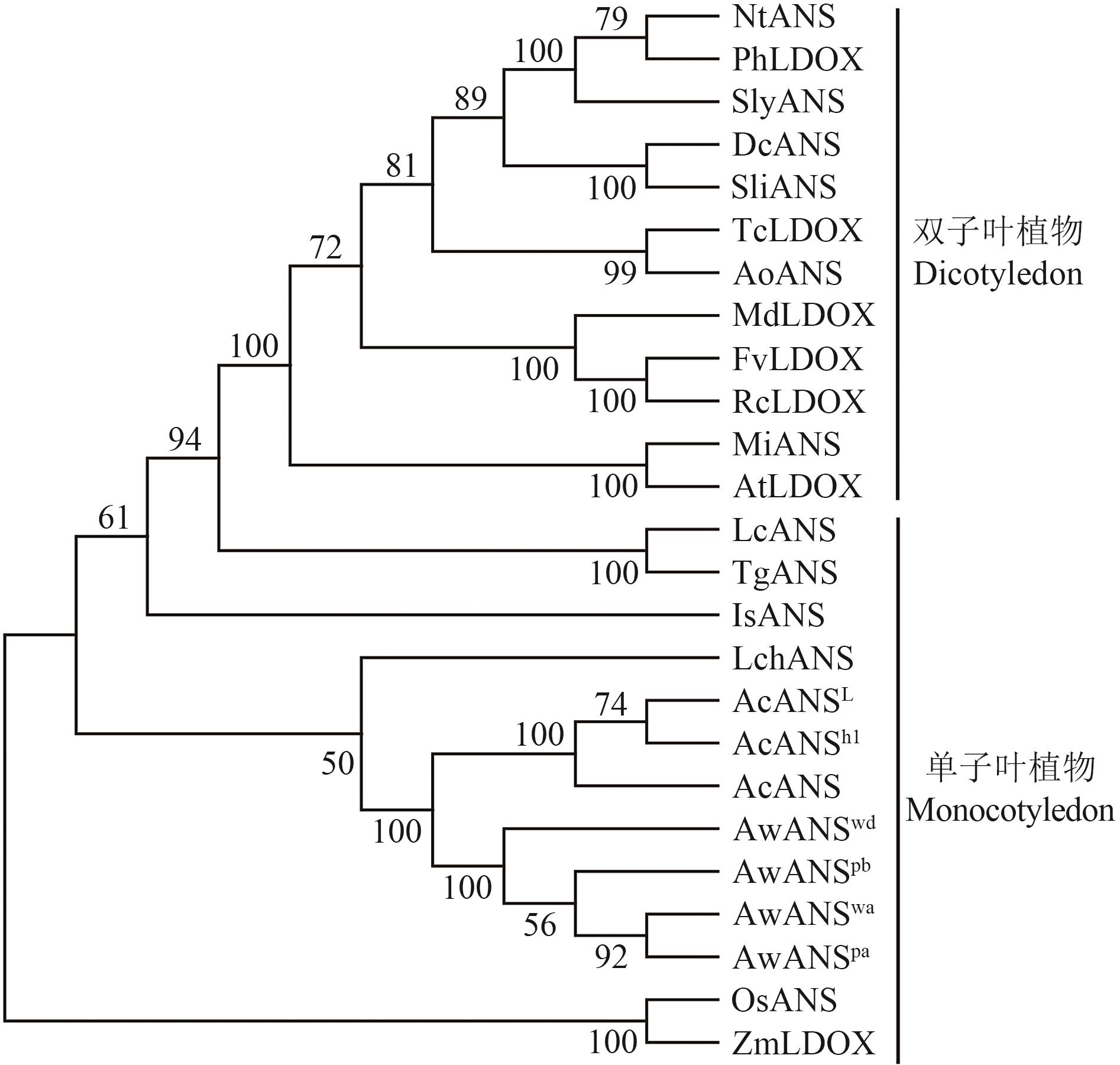
图5 多星韭AwANSs与其他物种ANS的系统进化树Nt:烟草;Ph:矮牵牛;Sl:番茄;Dc:香石竹;Sli:滨雪轮;Tc:可可;Ao:药葵;Md:苹果;Fv:野草莓;Rc:中国月季;Mi:紫罗兰;At:拟南芥;Lc:垂花百合;Tg:郁金香;Is:溪荪;Os:水稻;Zm:玉米;Lch:中国石蒜;Ac:洋葱;Aw:多星韭
Fig. 5 Phylogenetic analysis of AwANSs from A. wallichii and ANS from other speciesNt: Nicotiana tabacum; Ph: Petunia×hybrida; Sly: Solanum lycopersicum; Dc: Dianthus caryophyllus; Sli: Silene littorea; Tc: Theobroma cacao; Ao: Althaea officinalis; Md: Malus domestica; Fv: Fragaria vesca subsp. Vesca; Rc: Rosa chinensis; Mi: Matthiola incana; At: Arabidopsis thaliana; Lc: Lilium cernuum; Tg: Tulipa gesneriana; Is: Iris sanguinea; Os: Oryza sativa; Zm: Zea mays; Lch: Lycoris chinensis; Ac: Allium cepa; Aw: Allium wallichii
| 1 | Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids [J]. Plant J, 2008, 54(4): 733-749. |
| 2 | Zhao XC, Zhang YR, Long T, et al. Regulation mechanism of plant pigments biosynthesis: anthocyanins, carotenoids, and betalains [J]. Metabolites, 2022, 12(9): 871. |
| 3 | 刘玥, 李月庆, 孟祥宇, 等. 大花君子兰查尔酮合酶基因CmCHS的克隆及其功能验证 [J]. 园艺学报, 2021, 48(10): 1847-1858. |
| Liu Y, Li YQ, Meng XY, et al. Cloning and functional characterization of chalcone synthase genes (CmCHS) from Clivia miniata [J]. Acta horticulturae sinica, 2021, 48(10): 1847-1858. | |
| 4 | Li XL, Fan JZ, Luo SM, et al. Comparative transcriptome analysis identified important genes and regulatory pathways for flower color variation in Paphiopedilum hirsutissimum [J]. BMC Plant Biol, 2021, 21(1): 495. |
| 5 | Gu ZY, Zhu J, Hao Q, et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa) [J]. Plant Cell Physiol, 2019, 60(3): 599-611. |
| 6 | Wang S, Li LX, Zhang Z, et al. Ethylene precisely regulates anthocyanin synthesis in apple via a module comprising MdEIL1, MdMYB1, and MdMYB17 [J]. Hortic Res, 2022, 9: uhac034. |
| 7 | Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits [J]. Trends Plant Sci, 2013, 18(9): 477-483. |
| 8 | 赵维萍, 丁子俊, 王方平, 等. ‘滁菊’花青素合成酶(CmANS)基因的分子特征、原核表达与表达分析 [J]. 植物生理学报, 2022, 58(4): 677-686. |
| Zhao WP, Ding ZJ, Wang FP, et al. Molecular characterization, prokaryotic expression and expression pattern analysis of an anthocyanin synthase gene from Chrysanthemum morifolium ‘Chuju’[J]. Plant Physiol J, 2022, 58(4): 677-686. | |
| 9 | Feng YQ, Tian XC, Liang W, et al. Genome-wide identification of grape ANS gene family and expression analysis at different fruit coloration stages [J]. BMC Plant Biol, 2023, 23(1): 632. |
| 10 | 黄玲, 胡先梅, 梁泽慧, 等. 郁金香花青素合成酶基因TgANS的克隆与功能鉴定 [J]. 园艺学报, 2022, 49(9): 1935-1944. |
| Huang L, Hu XM, Liang ZH, et al. Cloning and function identification of anthocyanidin synthase gene TgANS in Tulipa gesneriana [J]. Acta Hortic Sin, 2022, 49(9): 1935-1944. | |
| 11 | Wu XX, Zhou Y, Yao D, et al. DNA methylation of LDOX gene contributes to the floral colour variegation in peach [J]. J Plant Physiol, 2020, 246/247: 153116. |
| 12 | Zhang HL, Zhao XJ, Zhang JP, et al. Functional analysis of an anthocyanin synthase gene StANS in potato [J]. Sci Hortic, 2020, 272: 109569. |
| 13 | Weiss D, van der Luit AH, Kroon JT, et al. The Petunia homologue of the Antirrhinum majus candi and Zea mays A2 flavonoid genes; homology to flavanone 3-hydroxylase and ethylene-forming enzyme [J]. Plant Mol Biol, 1993, 22(5): 893-897. |
| 14 | Nakajima J, Tanaka Y, Yamazaki M, et al. Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis [J]. J Biol Chem, 2001, 276(28): 25797-25803. |
| 15 | Szankowski I, Flachowsky H, Li HH, et al. Shift in polyphenol profile and sublethal phenotype caused by silencing of anthocyanidin synthase in apple (Malus sp.) [J]. Planta, 2009, 229(3): 681-692. |
| 16 | Lin CC, Xing PY, Jin H, et al. Loss of anthocyanidin synthase gene is associated with white flowers of Salvia miltiorrhiza Bge. f. alba, a natural variant of S. miltiorrhiza [J]. Planta, 2022, 256(1): 15. |
| 17 | Ben-Simhon Z, Judeinstein S, Trainin T, et al. A"white" anthocyanin-less pomegranate (Punica granatum L.) caused by an insertion in the coding region of the leucoanthocyanidin dioxygenase (LDOX; ANS) gene [J]. PLoS One, 2015, 10(11): e0142777. |
| 18 | Chen X, Krug L, Yang MF, et al. The Himalayan onion (Allium wallichii kunth) harbors unique spatially organized bacterial communities [J]. Microb Ecol, 2021, 82(4): 909-918. |
| 19 | 霍冬敖, 田瑞丰, 任永权, 等. 基于UPLC-MS/MS技术的野生及栽培韭菜籽的代谢组学研究 [J]. 广西植物, 2022, 42(12): 1995-2006. |
| Huo DA, Tian RF, Ren YQ, et al. UPLC-MS/MS-based metabolomic characterization and contrastive analysis between Allium wallichii and A. tuberosum seeds [J]. Guihaia, 2022, 42(12): 1995-2006. | |
| 20 | Bhandari J, Muhammad B, Thapa P, et al. Study of phytochemical, anti-microbial, anti-oxidant, and anti-cancer properties of Allium wallichii [J]. BMC Complement Altern Med, 2017, 17(1): 102. |
| 21 | Iqbal J, Abbasi BA, Batool R, et al. Potential phytocompounds for developing breast cancer therapeutics: nature's healing touch [J]. Eur J Pharmacol, 2018, 827: 125-148. |
| 22 | 张宇澄, 周颂东, 任海燕, 等. 中国西南葱属10种20居群的核型研究 [J]. 武汉植物学研究, 2009, 27(4): 351-360. |
| Zhang YC, Zhou SD, Ren HY, et al. Karyotype in 20 populations belonging to 10 species of Allium from Southwest China [J]. J Wuhan Bot Res, 2009, 27(4): 351-360. | |
| 23 | 唐汉青, 王晓月, 张子楠, 等. 多星韭(Allium wallichii)的传粉生态学初探 [J]. 广西植物, 2020, 40(11): 1613-1622. |
| Tang HQ, Wang XY, Zhang ZN, et al. Pollination biology of Allium wallichii [J]. Guihaia, 2020, 40(11): 1613-1622. | |
| 24 | Lin YX, Hou GY, Jiang YY, et al. Joint transcriptomic and metabolomic analysis reveals differential flavonoid biosynthesis in a high-flavonoid strawberry mutant [J]. Front Plant Sci, 2022, 13: 919619. |
| 25 | Lin Y, Liu GF, Rao Y, et al. Identification and validation of reference genes for qRT-PCR analyses under different experimental conditions in Allium wallichii [J]. J Plant Physiol, 2023, 281: 153925. |
| 26 | Zhang SS, Lu SJ, Yi SS, et al. Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia [J]. Planta, 2017, 245(2): 439-457. |
| 27 | 王青, 吕彤, 吕英民. 百合花被片的结构和色素成分对花色的影响 [J]. 园艺学报, 2021, 48(10): 1873-1884. |
| Wang Q, Lü T, Lü YM. The impact of tepal structure and pigment composition on the flower colour of lily [J]. Acta Hortic Sin, 2021, 48(10): 1873-1884. | |
| 28 | Morita Y, Hoshino A. Recent advances in flower color variation and patterning of Japanese morning glory and Petunia [J]. Breed Sci, 2018, 68(1): 128-138. |
| 29 | 邹晓菊, 黄瑞复, 翟书华. 六倍体多星韭的发现及多星韭种内倍性组成及演化的分析 [J]. 西北植物学报, 2013, 33(6): 1114-1122. |
| Zou XJ, Huang RF, Zhai SH. Detection of hexaploid and mechanisms of polyploidy formation in Allium wallichii kunth [J]. Acta Bot Boreali Occidentalia Sin, 2013, 33(6): 1114-1122. | |
| 30 | 赵莉丽, 王锦. 昆明梁王山多星韭与滇韭的核型比较 [J]. 西南林学院学报, 2009, 29(3): 22-25. |
| Zhao LL, Wang J. Comparative analysis on karyotype of Allium wallichii and Allium mairei from mount Liangwang in Kunming area [J]. J Southwest For Coll, 2009, 29(3): 22-25. | |
| 31 | 杨爽, 王锦. 云南多星韭细胞核型研究 [J]. 西南林学院学报, 2007, 27(5): 38-41. |
| Yang S, Wang J. Study on cytological karyotype of Allium wallichii in Liangwang Mountain area, Yunnan Province [J]. J Southwest For Coll, 2007, 27(5): 38-41. | |
| 32 | 翟艳红, 杨莹, 星耀武, 等. 云贵高原多星韭二倍体—四倍体分布格局研究 [J]. 植物科学学报, 2011, 29(1): 50-57. |
| Zhai YH, Yang Y, Xing YW, et al. Diploid and tetraploid distribution of Allium wallichii Kunth (Ailiaceae) in the Yunnan-Guizhou Plateau [J]. Plant Sci J, 2011, 29(1): 50-57. | |
| 33 | 安维忠, 刘雅莉, 刘红利, 等. 葡萄风信子MaANS基因及启动子的克隆与分析 [J]. 西北植物学报, 2015, 35(9): 1728-1734. |
| An WZ, Liu YL, Liu HL, et al. Isolation and analysis of Muscari armeniacum MaANS gene and its promoter [J]. Acta Bot Boreali Occidentalia Sin, 2015, 35(9): 1728-1734. | |
| 34 | Nakatsuka T, Nishihara M, Mishiba K, et al. Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants [J]. Plant Sci, 2005, 168(5): 1309-1318. |
| 35 | Rosati C, Cadic A, Duron M, et al. Molecular characterization of the anthocyanidin synthase gene in Forsythia × intermedia reveals organ-specific expression during flower development [J]. Plant Sci, 1999, 149(1): 73-79. |
| 36 | 洪艳, 武宇薇, 宋想, 等. 光照调控园艺作物花青素苷生物合成的分子机制 [J]. 园艺学报, 2021, 48(10): 1983-2000. |
| Hong Y, Wu YW, Song X, et al. Molecular mechanism of light-induced anthocyanin biosynthesis in horticultural crops [J]. Acta Hortic Sin, 2021, 48(10): 1983-2000. | |
| 37 | Grotewold E. The genetics and biochemistry of floral pigments [J]. Annu Rev Plant Biol, 2006, 57: 761-780. |
| 38 | Davies KM, Albert NW, Schwinn KE. From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning [J]. Funct Plant Biol, 2012, 39(8): 619-638. |
| 39 | 刘恺媛, 王茂良, 辛海波, 等. 植物花青素合成与调控研究进展 [J]. 中国农学通报, 2021, 37(14): 41-51. |
| Liu KY, Wang ML, Xin HB, et al. Anthocyanin biosynthesis and regulate mechanisms in plants: a review [J]. Chin Agric Sci Bull, 2021, 37(14): 41-51. | |
| 40 | 郑清冬, 王艺, 欧悦, 等. 兰科植物花色相关基因研究进展 [J]. 园艺学报, 2021, 48(10): 2057-2072. |
| Zheng QD, Wang Y, Ou Y, et al. Research advances of genes responsible for flower colors in Orchidaceae [J]. Acta Hortic Sin, 2021, 48(10): 2057-2072. | |
| 41 | Nakatsuka T, Saito M, Sato-Ushiku Y, et al. Development of DNA markers that discriminate between white- and blue-flowers in Japanese gentian plants [J]. Euphytica, 2012, 184(3): 335-344. |
| 42 | Rafique MZ, Carvalho E, Stracke R, et al. Nonsense mutation inside anthocyanidin synthase gene controls pigmentation in yellow raspberry (Rubus idaeus L.) [J]. Front Plant Sci, 2016, 7: 1892. |
| 43 | 王晰, 丁文杰, 李娅, 等. 长筒石蒜花青素合成酶基因LlANS的克隆与表达分析 [J]. 河南农业大学学报, 2018, 52(4): 611-617. |
| Wang X, Ding WJ, Li Y, et al. Cloning and expression analysis of anthocyanidin synthase(LlANS) in Lycoris longituba [J]. J Henan Agric Univ, 2018, 52(4): 611-617. | |
| 44 | Oren-Shamir M. Does anthocyanin degradation play a significant role in determining pigment concentration in plants? [J]. Plant Sci, 2009, 177(4): 310-316. |
| 45 | Luo HH, Deng SF, Fu W, et al. Characterization of active anthocyanin degradation in the petals of Rosa chinensis and Brunfelsia calycina reveals the effect of gallated catechins on pigment maintenance [J]. Int J Mol Sci, 2017, 18(4): 699. |
| 46 | Zipor G, Duarte P, Carqueijeiro I, et al. In planta anthocyanin degradation by a vacuolar class Ⅲ peroxidase in Brunfelsia calycina flowers [J]. New Phytol, 2015, 205(2): 653-665. |
| 47 | Liu J, Wang YX, Zhang MH, et al. Color fading in Lotus (Nelumbo nucifera) petals is manipulated both by anthocyanin biosynthesis reduction and active degradation [J]. Plant Physiol Biochem, 2022, 179: 100-107. |
| 48 | Li SH, He YJ, Li LZ, et al. New insights on the regulation of anthocyanin biosynthesis in purple Solanaceous fruit vegetables [J]. Sci Hortic, 2022, 297: 110917. |
| [1] | 许圆梦, 毛娇, 王梦瑶, 王数, 任江陵, 刘宇涵, 刘思辰, 乔治军, 王瑞云, 曹晓宁. 糜子PmDEP1和PmEP3基因的克隆与表达特征分析[J]. 生物技术通报, 2025, 41(2): 150-162. |
| [2] | 焦小雨, 吴琼, 刘丹丹, 孙明慧, 阮旭, 王雷刚, 王文杰. 茶树CsWAK8克隆及其在响应冷胁迫过程中的功能分析[J]. 生物技术通报, 2025, 41(2): 210-220. |
| [3] | 乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103. |
| [4] | 庞梦真, 徐汉琴, 刘海燕, 宋娟, 王佳涵, 孙丽娜, 姬佩梅, 尹泽芝, 胡又川, 赵晓萌, 梁闪闪, 张泗举, 栾维江. 水稻黄化早抽穗突变体 hz1 的基因鉴定及功能分析[J]. 生物技术通报, 2024, 40(7): 125-136. |
| [5] | 沈真辉, 曹瑶, 杨林雷, 罗祥英, 子灵山, 陆青青, 李荣春. 金耳和毛韧革菌麦角硫因生物合成基因的克隆及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 259-272. |
| [6] | 黄丹, 姜山, 彭涛. 褐角苔FfCYP98基因克隆及其功能分析[J]. 生物技术通报, 2024, 40(7): 273-284. |
| [7] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [8] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [9] | 杜泽光, 任少文, 张凤勤, 李梅兰, 李改珍, 齐仙惠. 大白菜BrMLP328的克隆、表达及功能验证[J]. 生物技术通报, 2024, 40(4): 122-129. |
| [10] | 刘换换, 杨立春, 李火根. 北美鹅掌楸LtMYB305基因的克隆及功能分析[J]. 生物技术通报, 2024, 40(4): 179-188. |
| [11] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [12] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [13] | 苑馨予, 钟彩虹, 张龙, 郑浩, 李吉涛, 张琼. 猕猴桃AcMYB88的鉴定及功能研究[J]. 生物技术通报, 2024, 40(2): 183-196. |
| [14] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [15] | 华炫, 田博雯, 周欣彤, 江梓涵, 王诗琦, 黄倩慧, 张健, 陈艳红. 旱柳SmERF B3-45的克隆及耐盐功能研究[J]. 生物技术通报, 2024, 40(12): 124-135. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||