








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 166-175.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0878
• 研究报告 • 上一篇
王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静( )
)
收稿日期:2024-09-09
出版日期:2025-04-26
发布日期:2025-04-25
通讯作者:
王静,女,博士,讲师,研究方向 :辣椒功能基因挖掘与种质创新;E-mail: wangjing315@sxau.edu.cn基金资助:
WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing( )
)
Received:2024-09-09
Published:2025-04-26
Online:2025-04-25
摘要:
目的 研究辣椒(Capsicum annuum L.)GASA的基因结构和表达分析,为阐明CaGASA基因的生物学功能及进一步培育抗逆性强的辣椒品种提供参考。 方法 运用生物信息学方法对辣椒8个GASA进行鉴定,并分析它们的理化性质、系统发育关系、基因结构、保守基序、染色体位置、蛋白互作网络、基因同源性和启动子区域等,实时荧光定量PCR(real-time fluorescence quantitative PCR, RT-qPCR)验证辣椒GASA基因家族成员在不同组织、非生物胁迫和激素处理下的表达模式。 结果 8个CaGASA在辣椒5条染色体上不均匀分布,所有CaGASA蛋白都有一个共同的结构域GASA;亚细胞定位预测结果表明,CaGASA蛋白存在于细胞外区域;辣椒、拟南芥(Arabidopsis thaliana)、番茄(Solanum lycopersicum L.)、水稻(Oryza sativa L.)基因组的直系同源GASA基因共线性分析结果表明,辣椒与其他3个物种之间有14个共线性事件,CaGASA5与3个物种的GASA基因均具有共线性;顺式元件分析表明,GASA基因可能受到多种植物激素和胁迫的诱导;转录组数据分析显示,CaGASA在辣椒花和果实中的表达水平高于叶;RT-qPCR分析表明,CaGASA基因响应低温、高温胁迫和脱落酸(abscisic acid, ABA)、赤霉素(gibberellin, GA)、吲哚乙酸(indole acetic acid, IAA)和茉莉酸(jasmonic acid, MeJA)激素处理。 结论 辣椒GASA基因家族可能参与辣椒花发育,响应激素和耐高、低温胁迫的应答。
王田田, 常雪瑞, 黄婉洋, 黄嘉欣, 苗如意, 梁燕平, 王静. 辣椒GASA基因家族的鉴定及分析[J]. 生物技术通报, 2025, 41(4): 166-175.
WANG Tian-tian, CHANG Xue-rui, HUANG Wan-yang, HUANG Jia-xin, MIAO Ru-yi, LIANG Yan-ping, WANG Jing. Identification and Analysis of GASA Gene Family in Pepper (Capsicum annuum L.)[J]. Biotechnology Bulletin, 2025, 41(4): 166-175.
| 基因 Gene | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) |
|---|---|---|
| CaGASA1 | CATTCCAAAAGGCATTTGCT | GCAAGGGCATGCTTCATAAT |
| CaGASA2 | TGCAGCTTTGCTTATTGCAT | GTACGCAGTGGCATCTGTTG |
| CaGASA3 | GCAGCAGCAACAACAAAGAA | TAGCAAGGGCAAGTTTGCTT |
| CaGASA4 | TGATGCCCAGTACCACATTG | GGTCCTCCTTCCTTGGTCTT |
| CaGASA5 | ACTTGAACAACGGGGATTTG | AAGGGCACTTGGTCTTGTTG |
| CaGASA6 | CCACTGCACCGACATACATT | TGGTGGAACACATTGACACC |
| CaGASA7 | TCTCTCCTTGCTCCTTCTCG | GCAAGGGCAAGTCTGAGTGT |
| CaGASA8 | TGAGCTTGTGGAAGCTGATG | CACCATGGGTAAGCATGTCA |
| β-Actin | CAACATCTTCACTCTCTGCTCT | ACTAGGAAAAACAGCACTTGGT |
表1 CaGASA家族基因表达分析的RT-qPCR引物
Table 1 RT-qPCR primers for expression analysis of CaGASA family genes
| 基因 Gene | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) |
|---|---|---|
| CaGASA1 | CATTCCAAAAGGCATTTGCT | GCAAGGGCATGCTTCATAAT |
| CaGASA2 | TGCAGCTTTGCTTATTGCAT | GTACGCAGTGGCATCTGTTG |
| CaGASA3 | GCAGCAGCAACAACAAAGAA | TAGCAAGGGCAAGTTTGCTT |
| CaGASA4 | TGATGCCCAGTACCACATTG | GGTCCTCCTTCCTTGGTCTT |
| CaGASA5 | ACTTGAACAACGGGGATTTG | AAGGGCACTTGGTCTTGTTG |
| CaGASA6 | CCACTGCACCGACATACATT | TGGTGGAACACATTGACACC |
| CaGASA7 | TCTCTCCTTGCTCCTTCTCG | GCAAGGGCAAGTCTGAGTGT |
| CaGASA8 | TGAGCTTGTGGAAGCTGATG | CACCATGGGTAAGCATGTCA |
| β-Actin | CAACATCTTCACTCTCTGCTCT | ACTAGGAAAAACAGCACTTGGT |
基因号 Gene ID | 基因 Gene | 氨基酸数 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点 Theoretical pI | 不稳定指数 Instability index | 脂肪指数 Aliphatic index | 总平均亲水系数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization | 跨膜结构域 Transmembrane domain |
|---|---|---|---|---|---|---|---|---|---|
| Capana02g002587 | CaGASA1 | 102 | 11.15 | 9.01 | 46.06 | 49.90 | -0.288 | 细胞外 | 0 |
| Capana02g002588 | CaGASA2 | 101 | 10.97 | 9.12 | 72.87 | 56.14 | -0.166 | 细胞外 | 1 |
| Capana02g003203 | CaGASA3 | 112 | 12.67 | 9.42 | 42.06 | 53.04 | -0.478 | 细胞外 | 0 |
| Capana03g000783 | CaGASA4 | 104 | 11.46 | 9.35 | 45.52 | 62.88 | -0.026 | 细胞外 | 1 |
| Capana03g001010 | CaGASA5 | 145 | 15.74 | 9.27 | 39.20 | 81.38 | -0.043 | 细胞外 | 0 |
| Capana06g002956 | CaGASA6 | 113 | 12.55 | 9.46 | 48.91 | 73.27 | 0.103 | 细胞外 | 1 |
| Capana08g002677 | CaGASA7 | 104 | 11.18 | 9.24 | 37.42 | 71.44 | -0.138 | 细胞外 | 0 |
| Capana12g000591 | CaGASA8 | 113 | 12.27 | 7.97 | 36.80 | 79.38 | 0.040 | 细胞外 | 0 |
表2 辣椒GASA基因家族成员信息
Table 2 Information of GASA gene family members in pepper
基因号 Gene ID | 基因 Gene | 氨基酸数 Number of amino acids/aa | 分子量 Molecular weight/kD | 等电点 Theoretical pI | 不稳定指数 Instability index | 脂肪指数 Aliphatic index | 总平均亲水系数 Grand average of hydropathicity | 亚细胞定位 Subcellular localization | 跨膜结构域 Transmembrane domain |
|---|---|---|---|---|---|---|---|---|---|
| Capana02g002587 | CaGASA1 | 102 | 11.15 | 9.01 | 46.06 | 49.90 | -0.288 | 细胞外 | 0 |
| Capana02g002588 | CaGASA2 | 101 | 10.97 | 9.12 | 72.87 | 56.14 | -0.166 | 细胞外 | 1 |
| Capana02g003203 | CaGASA3 | 112 | 12.67 | 9.42 | 42.06 | 53.04 | -0.478 | 细胞外 | 0 |
| Capana03g000783 | CaGASA4 | 104 | 11.46 | 9.35 | 45.52 | 62.88 | -0.026 | 细胞外 | 1 |
| Capana03g001010 | CaGASA5 | 145 | 15.74 | 9.27 | 39.20 | 81.38 | -0.043 | 细胞外 | 0 |
| Capana06g002956 | CaGASA6 | 113 | 12.55 | 9.46 | 48.91 | 73.27 | 0.103 | 细胞外 | 1 |
| Capana08g002677 | CaGASA7 | 104 | 11.18 | 9.24 | 37.42 | 71.44 | -0.138 | 细胞外 | 0 |
| Capana12g000591 | CaGASA8 | 113 | 12.27 | 7.97 | 36.80 | 79.38 | 0.040 | 细胞外 | 0 |
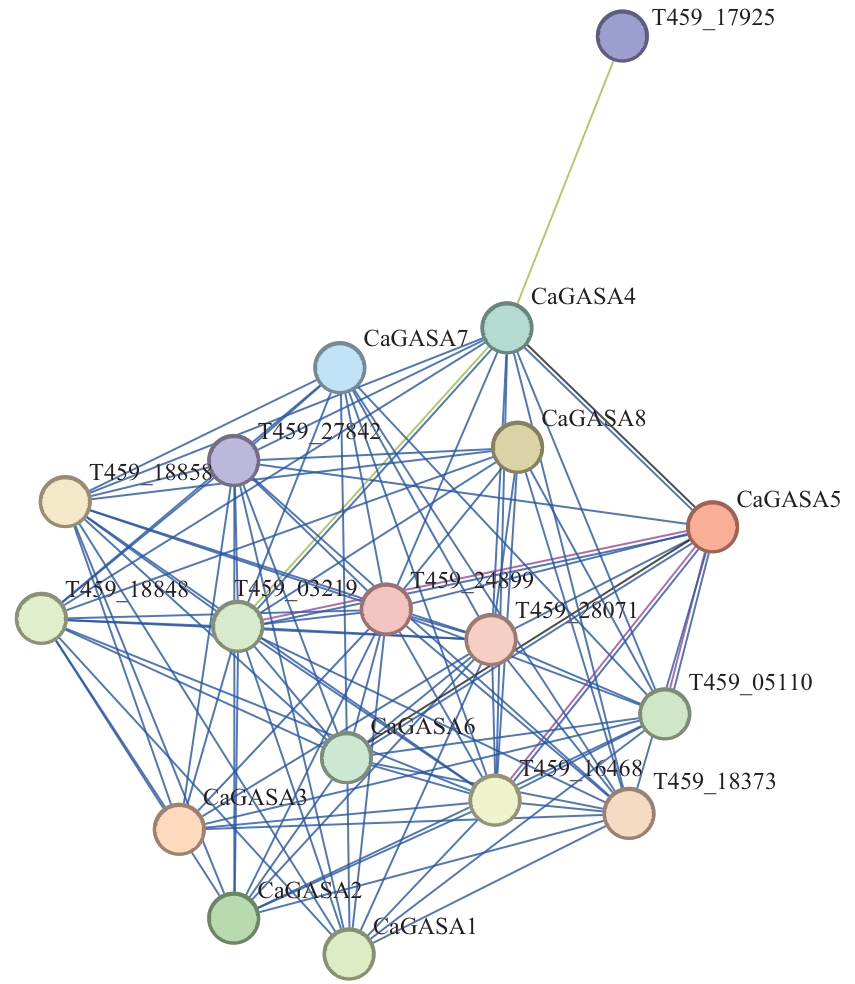
图3 CaGASA蛋白互作网络分析图T459_27842:含l9抑制子蛋白域蛋白;T459_18858:假定的嘌呤通透酶5;T459_18848:假定的嘌呤通透酶5;T459_03219:含Fe2OG双加氧酶结构域蛋白;T459_17925、T459_24899、T459_28071、T459_18373、T459_16468、T459_05110:未鉴定的蛋白。紫线表示实验测定;蓝线表示共同产生;黄线表示文本挖掘证据;黑线表示共表达
Fig. 3 Interaction network analysis of CaGASA proteinsT459_27842:Inhibitor I9 domain-containing protein. T459_18858: Putative purine permease 5. T459_18848: Putative purine permease 5. T459_03219: Fe2OG dioxygenase domain-containing protein. T459_17925, T459_24899, T459_28071, T459_18373, T459_16468, T459_05110: Uncharacterized protein. Purple line indicates experimental determination; the blue line indicates co-occurrence; the yellow line indicates text mining evidence; and the black line indicates co-expression
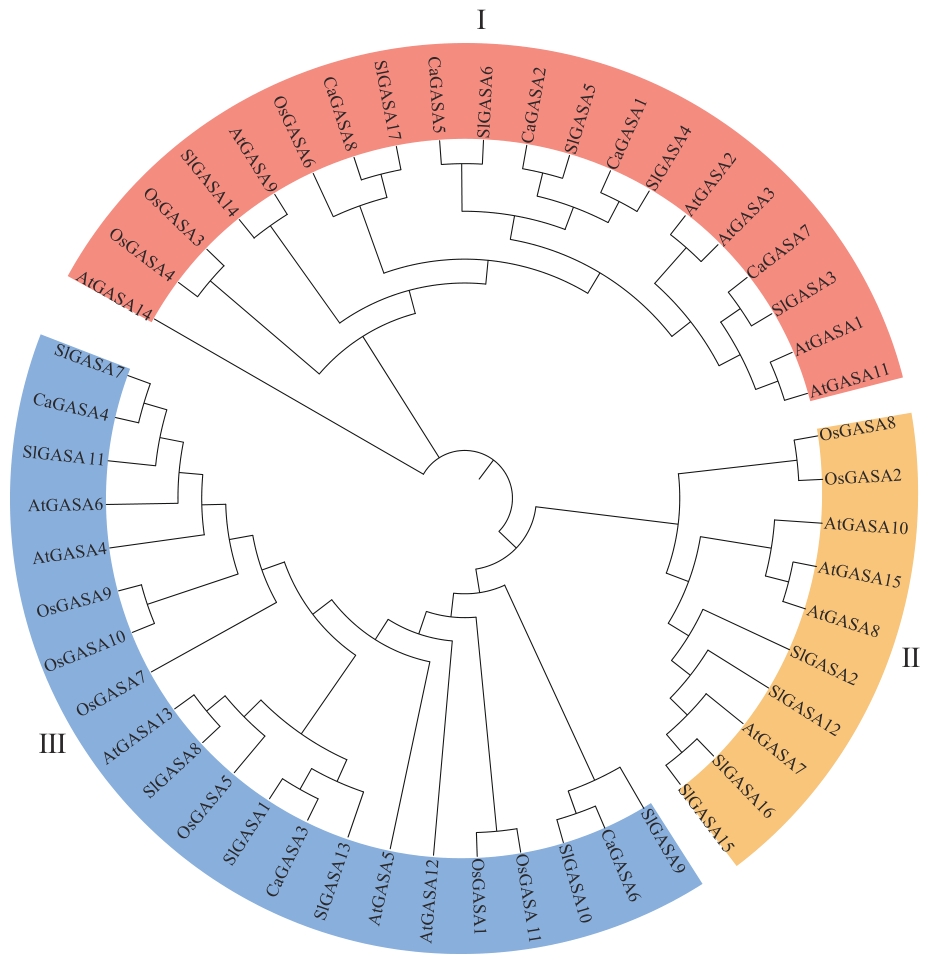
图4 辣椒(Ca)和拟南芥(At)、番茄(Sl)、水稻(Os)GASA家族成员系统发育树
Fig. 4 Phylogenetic tree of GASA family members of Arabidopsis thaliana (At), Solanum lycopersicum (Sl), Oryza sativa (Os), and Capsicum annuum (Ca)
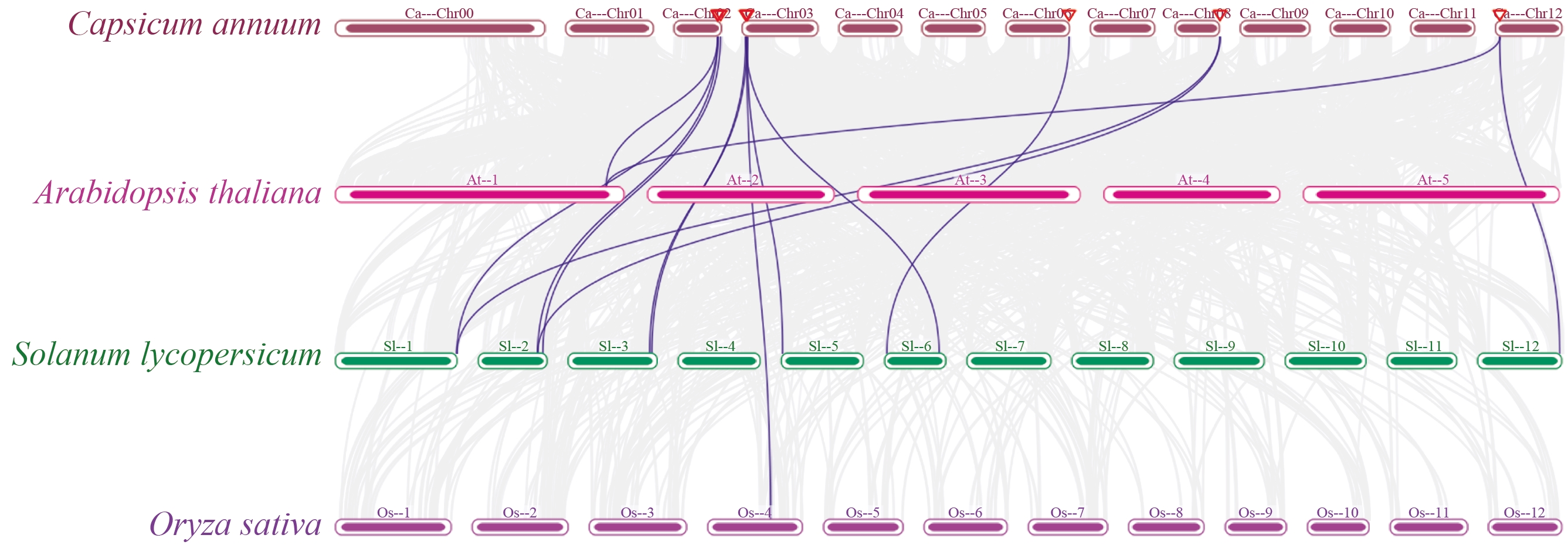
图6 辣椒(Ca)和拟南芥(At)、番茄(Sl)、水稻(Os)GASA基因的同源分析
Fig. 6 Synteny analysis of GASA genes among C. annuum (Ca), A. thaliana (At), S. lycopersicum (Sl), and O. sativa (Os)

图8 辣椒GASA家族基因在不同组织中的表达分析L1‒L9代表新叶刚出现后2、5、10、15、20、30、40、50和60 d;F1‒F9代表出现花蕾后;G1‒G11代表授粉后10、15、20、25、30、35、40、45、50、55和60 d;ST1、ST2代表授粉后10和15 d;S3‒S11代表授粉后20、25、30、35、40、45、50、55和60 d;T3‒T11代表授粉后20、25、30、35、40、45、50、55和60 d
Fig. 8 Expression analysis of GASA in different tissues in pepperL1‒L9 refers to 2, 5, 10, 15, 20, 30, 40, 50, and 60 d after new leaf emergence; F1‒F9 refers to 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, and 60 d after flower bud emergence; G1‒G11 refers to 10, 15, 20, 25 30, 35, 40, 45, 50, 55, and 60 d after pollination; St1 and St2 refers to 10, 15 d after pollination; S3‒S11 refer to 20, 25, 30, 35, 40, 45, 50, 55, and 60 d after pollination; T3‒T11 refers to 20, 25, 30, 35, 40, 45, 50, 55, and 60 d after pollination
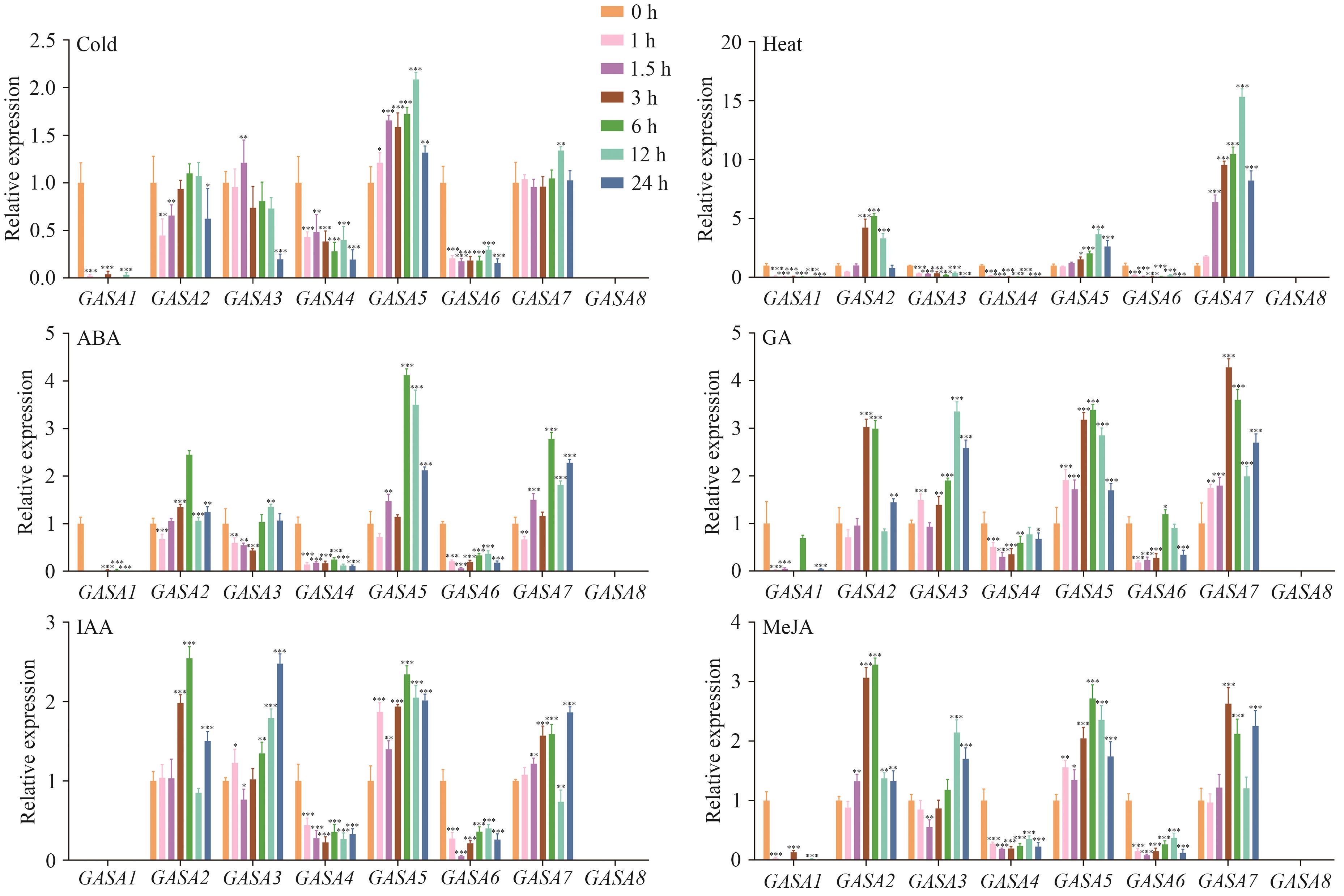
图9 八个CaGASA基因在非生物胁迫处理下的相对表达量*、**、***分别表示在0.05、0.01、0.001水平上差异显著
Fig. 9 Relative expressions of eight CaGASA genes under abiotic stress treatment*, **, and *** indicate significant differences at the level of 0.05, 0.01 and 0.001 respectively
| 1 | 邹学校, 朱凡. 辣椒的起源、进化与栽培历史 [J]. 园艺学报, 2022, 49(6): 1371-1381. |
| Zou XX, Zhu F. Origin, evolution and cultivation history of the pepper [J]. Acta Hortic Sin, 2022, 49(6): 1371-1381. | |
| 2 | 邓惠如, 张素勤, 耿广东. 辣椒逆境响应基因研究现状及前景 [J/OL]. 分子植物育种, 2023. . |
| Deng HR, Zhang SQ, Geng GD. Research status and prospect of stress response genes in pepper [J/OL]. China Ind Econ, 2023. . | |
| 3 | Bouteraa MT, Ben Romdhane W, Baazaoui N, et al. GASA proteins: review of their functions in plant environmental stress tolerance [J]. Plants, 2023, 12(10): 2045. |
| 4 | Taylor BH, Scheuring CF. A molecular marker for lateral root initiation: the RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia [J]. Mol Gen Genet, 1994, 243(2): 148-157. |
| 5 | Herzog M, Dorne AM, Grellet F. GASA a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene [J]. Plant Mol Biol, 1995, 27(4): 743-752. |
| 6 | Su D, Liu KD, Yu ZS, et al. Genome-wide characterization of the tomato GASA family identifies SlGASA1 as a repressor of fruit ripening [J]. Hortic Res, 2023, 10(1): uhac222. |
| 7 | Segura A, Moreno M, Madueño F, et al. Snakin-1, a peptide from potato that is active against plant pathogens [J]. Mol Plant Microbe Interact, 1999, 12(1): 16-23. |
| 8 | Wigoda N, Ben-Nissan G, Granot D, et al. The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity [J]. Plant J, 2006, 48(5): 796-805. |
| 9 | Wang L, Wang Z, Xu YY, et al. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice [J]. Plant J, 2009, 57(3): 498-510. |
| 10 | Cheng XR, Wang SX, Xu DM, et al. Identification and analysis of the GASR gene family in common wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination [J]. Front Genet, 2019, 10: 980. |
| 11 | Peng JZ, Lai LJ, Wang XJ. PRGL: a cell wall proline-rich protein containning GASA domain in Gerbera hybrida [J]. Sci China C Life Sci, 2008, 51(6): 520-525. |
| 12 | Liu ZH, Zhu L, Shi HY, et al. Cotton GASL genes encoding putative gibberellin-regulated proteins are involved in response to GA signaling in fiber development [J]. Mol Biol Rep, 2013, 40(7): 4561-4570. |
| 13 | Zhong CM, Xu H, Ye ST, et al. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis [J]. Plant Physiol, 2015, 169(3): 2288-2303. |
| 14 | Zimmermann R, Sakai H, Hochholdinger F. The Gibberellic Acid Stimulated-Like gene family in maize and its role in lateral root development [J]. Plant Physiol, 2010, 152(1): 356-365. |
| 15 | Zhang SC, Yang CW, Peng JZ, et al. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana [J]. Plant Mol Biol, 2009, 69(6): 745-759. |
| 16 | Li Y, Yuan WM, Peng JL, et al. GhGASA14 regulates the flowering time of upland cotton in response to GA3 [J]. Plant Cell Rep, 2024, 43(7): 170. |
| 17 | He ZY, Jiang R, Wang XJ, et al. A GASA protein family gene, CmGEG, inhibits petal growth in Chrysanthemum [J]. Int J Mol Sci, 2024, 25(6): 3367. |
| 18 | Syed Nabi RB, Lee MH, Cho KS, et al. Genome-wide identification and comprehensive analysis of the GASA gene family in peanuts (Arachis hypogaea L.) under abiotic stress [J]. Int J Mol Sci, 2023, 24(23): 17117. |
| 19 | Iqbal A, Khan RS. Snakins: antimicrobial potential and prospects of genetic engineering for enhanced disease resistance in plants [J]. Mol Biol Rep, 2023, 50(10): 8683-8690. |
| 20 | Panji A, Ismaili A, Sohrabi SM. Genome-wide identification and expression profiling of snakin/GASA genes under drought stress in barley (Hordeum vulgare L.) [J]. 3 Biotech, 2023, 13(5): 126. |
| 21 | Lee SH, Yoon JS, Jung WJ, et al. Genome-wide identification and characterization of the lettuce GASA family in response to abiotic stresses [J]. BMC Plant Biol, 2023, 23(1): 106. |
| 22 | Chen BJ, Sun YR, Tian ZL, et al. GhGASA10-1 promotes the cell elongation in fiber development through the phytohormones IAA-induced [J]. BMC Plant Biol, 2021, 21(1): 448. |
| 23 | Rubinovich L, Ruthstein S, Weiss D. The Arabidopsis cysteine-rich GASA5 is a redox-active metalloprotein that suppresses gibberellin responses [J]. Mol Plant, 2014, 7(1): 244-247. |
| 24 | Qu J, Kang SG, Hah C, et al. Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana [J]. Plant Sci, 2016, 246: 1-10. |
| 25 | 马艳青, 戴雄泽, 李雪峰, 等. 辣椒骨干亲本6421及其衍生系的创制与利用 [J]. 中国蔬菜, 2015(6): 11-16. |
| Ma YQ, Dai XZ, Li XF, et al. Creation and utilization of pepper backbone parent 6421 and its derivated lines [J]. China Veg, 2015(6): 11-16. | |
| 26 | Li ZW, Gao JP, Wang GH, et al. Genome-wide identification and characterization of GASA gene family in Nicotiana tabacum [J]. Front Genet, 2022, 12: 768942. |
| 27 | Fan S, Zhang D, Zhang LZ, et al. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction [J]. BMC Genomics, 2017, 18(1): 827. |
| 28 | Yang MZ, Liu CY, Zhang W, et al. Genome-wide identification and characterization of gibberellic acid-stimulated Arabidopsis gene family in pineapple (Ananas comosus) [J]. Int J Mol Sci, 2023, 24(23): 17063. |
| 29 | Rezaee S, Ahmadizadeh M, Heidari P. Genome-wide characterization, expression profiling, and post-transcriptional study of GASA gene family [J]. Gene Rep, 2020, 20: 100795. |
| 30 | Guo RR, Xu XZ, Carole B, et al. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape [J]. BMC Genomics, 2013, 14: 554. |
| 31 | Chow CN, Lee TY, Hung YC, et al. PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants [J]. Nucleic Acids Res, 2019, 47(D1): D1155-D1163. |
| 32 | Ahmad B, Yao J, Zhang SL, et al. Genome-wide characterization and expression profiling of GASA genes during different stages of seed development in grapevine (Vitis vinifera L.) predict their involvement in seed development [J]. Int J Mol Sci, 2020, 21(3): 1088. |
| 33 | Ko CB, Woo YM, Lee DJ, et al. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene [J]. Plant Physiol Biochem, 2007, 45(9): 722-728. |
| 34 | Bouteraa MT, Ben Romdhane W, Ben Hsouna A, et al. Genome-wide characterization and expression profiling of GASA gene family in Triticum turgidum ssp. durum (desf.) husn. (Durum wheat) unveils its involvement in environmental stress responses [J]. Phytochemistry, 2023, 206: 113544. |
| 35 | Zhang LY, Geng XL, Zhang HY, et al. Isolation and characterization of heat-responsive gene TaGASR1 from wheat (Triticum aestivum L.) [J]. J Plant Biol, 2017, 60(1): 57-65. |
| 36 | Muhammad I, Li WQ, Jing XQ, et al. A systematic in silico prediction of gibberellic acid stimulated GASA family members: a novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice [J]. J Plant Physiol, 2019, 234/235: 117-132. |
| 37 | Li KL, Bai X, Li Y, et al. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs [J]. J Plant Physiol, 2011, 168(18): 2153-2160. |
| [1] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [2] | 孙天国, 衣兰, 秦旭洋, 乔梦雪, 谷新颖, 韩艺, 沙伟, 张梅娟, 马天意. 大白菜DABB基因家族的全基因组鉴定及盐碱胁迫下的表达分析[J]. 生物技术通报, 2025, 41(4): 156-165. |
| [3] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| [4] | 王琛, 刘国梅, 陈畅, 张晋龙, 姚琳, 孙璇, 杜春芳. 白菜型油菜CCDs家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(3): 161-170. |
| [5] | 彭婷, 林颖, 谭圆圆, 饶英, 黄覃, 张文娥, 汪波, 田瑞丰, 刘国锋. 多星韭AwANSs基因的克隆与表达分析[J]. 生物技术通报, 2025, 41(3): 230-239. |
| [6] | 马天意, 许家佳, 路文婧, 吴艳, 沙伟, 张梅娟, 彭疑芳. ‘金小童’大白菜BrcGASA3基因在盐碱胁迫下的表达分析及抗性鉴定[J]. 生物技术通报, 2025, 41(2): 127-138. |
| [7] | 许圆梦, 毛娇, 王梦瑶, 王数, 任江陵, 刘宇涵, 刘思辰, 乔治军, 王瑞云, 曹晓宁. 糜子PmDEP1和PmEP3基因的克隆与表达特征分析[J]. 生物技术通报, 2025, 41(2): 150-162. |
| [8] | 贾子健, 王宝强, 陈立飞, 王义真, 魏小红, 赵颖. 响应NO的藜麦CHX基因家族在盐碱胁迫下的表达模式[J]. 生物技术通报, 2025, 41(2): 163-174. |
| [9] | 钱政毅, 吴绍芳, 曹舒怡, 宋雅欣, 潘鑫峰, 李兆伟, 范凯. 睡莲NAC转录因子的鉴定及其表达分析[J]. 生物技术通报, 2025, 41(2): 234-247. |
| [10] | 黄颖, 遇文婧, 刘雪峰, 刁桂萍. 山新杨谷胱甘肽转移酶基因的生物信息学与表达模式分析[J]. 生物技术通报, 2025, 41(2): 248-256. |
| [11] | 向春繁, 李勒松, 王娟, 梁艳丽, 杨生超, 栗孟飞, 赵艳. 当归肉桂醇脱氢酶AsCAD功能鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 295-308. |
| [12] | 李明, 刘祥宇, 王益娜, 和四梅, 沙本才. 紫金龙异紫堇定生物合成相关6-OMT基因克隆与功能表征[J]. 生物技术通报, 2025, 41(2): 309-320. |
| [13] | 葛仕杰, 刘怡德, 张华东, 宁强, 朱展望, 王书平, 刘易科. 小麦蛋白质二硫键异构酶基因家族的鉴定与表达[J]. 生物技术通报, 2025, 41(2): 85-96. |
| [14] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [15] | 王子傲, 田瑞, 崔永梅, 白羿雄, 姚晓华, 安立昆, 吴昆仑. 青稞HvnJAZ4的生物信息学和表达模式分析[J]. 生物技术通报, 2025, 41(1): 173-185. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||